A quantitative atlas of mitotic phosphorylation
- PMID: 18669648
- PMCID: PMC2504835
- DOI: 10.1073/pnas.0805139105
A quantitative atlas of mitotic phosphorylation
Abstract
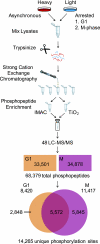
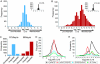
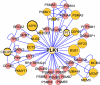
The eukaryotic cell division cycle is characterized by a sequence of orderly and highly regulated events resulting in the duplication and separation of all cellular material into two newly formed daughter cells. Protein phosphorylation by cyclin-dependent kinases (CDKs) drives this cycle. To gain further insight into how phosphorylation regulates the cell cycle, we sought to identify proteins whose phosphorylation is cell cycle regulated. Using stable isotope labeling along with a two-step strategy for phosphopeptide enrichment and high mass accuracy mass spectrometry, we examined protein phosphorylation in a human cell line arrested in the G(1) and mitotic phases of the cell cycle. We report the identification of >14,000 different phosphorylation events, more than half of which, to our knowledge, have not been described in the literature, along with relative quantitative data for the majority of these sites. We observed >1,000 proteins with increased phosphorylation in mitosis including many known cell cycle regulators. The majority of sites on regulated phosphopeptides lie in [S/T]P motifs, the minimum required sequence for CDKs, suggesting that many of the proteins may be CDK substrates. Analysis of non-proline site-containing phosphopeptides identified two unique motifs that suggest there are at least two undiscovered mitotic kinases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. - PubMed
-
- Massague J. G1 cell cycle control and cancer. Nature. 2004;432:298–306. - PubMed
-
- Rao PN, Johnson RT. Mammalian cell fusion: Studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. - PubMed
-
- Masui Y, Markert CL. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J Exp Zool. 1971;177:129–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

