Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies
- PMID: 18669659
- PMCID: PMC2504805
- DOI: 10.1073/pnas.0800658105
Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies
Abstract
Huntington's disease is a dominant autosomal neurodegenerative disorder caused by an expansion of polyglutamines in the huntingtin (Htt) protein, whose cellular function remains controversial. To gain insight into Htt function, we purified epitope-tagged Htt and identified Argonaute as associated proteins. Colocalization studies demonstrated Htt and Ago2 to be present in P bodies, and depletion of Htt showed compromised RNA-mediated gene silencing. Mouse striatal cells expressing mutant Htt showed fewer P bodies and reduced reporter gene silencing activity compared with wild-type counterparts. These data suggest that the previously reported transcriptional deregulation in HD may be attributed in part to mutant Htt's role in post-transcriptional processes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

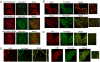
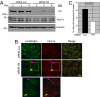


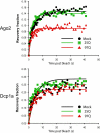
References
-
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. - PubMed
-
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. - PubMed
-
- Li SH, Li XJ. Huntingtin and its role in neuronal degeneration. Neuroscientist. 2004;10:467–475. - PubMed
-
- Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

