Gene networks driving bovine milk fat synthesis during the lactation cycle
- PMID: 18671863
- PMCID: PMC2547860
- DOI: 10.1186/1471-2164-9-366
Gene networks driving bovine milk fat synthesis during the lactation cycle
Abstract
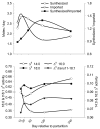
Background: The molecular events associated with regulation of milk fat synthesis in the bovine mammary gland remain largely unknown. Our objective was to study mammary tissue mRNA expression via quantitative PCR of 45 genes associated with lipid synthesis (triacylglycerol and phospholipids) and secretion from the late pre-partum/non-lactating period through the end of subsequent lactation. mRNA expression was coupled with milk fatty acid (FA) composition and calculated indexes of FA desaturation and de novo synthesis by the mammary gland.
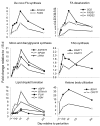
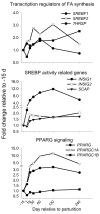
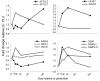
Results: Marked up-regulation and/or % relative mRNA abundance during lactation were observed for genes associated with mammary FA uptake from blood (LPL, CD36), intracellular FA trafficking (FABP3), long-chain (ACSL1) and short-chain (ACSS2) intracellular FA activation, de novo FA synthesis (ACACA, FASN), desaturation (SCD, FADS1), triacylglycerol synthesis (AGPAT6, GPAM, LPIN1), lipid droplet formation (BTN1A1, XDH), ketone body utilization (BDH1), and transcription regulation (INSIG1, PPARG, PPARGC1A). Change in SREBF1 mRNA expression during lactation, thought to be central for milk fat synthesis regulation, was < or =2-fold in magnitude, while expression of INSIG1, which negatively regulates SREBP activation, was >12-fold and had a parallel pattern of expression to PPARGC1A. Genes involved in phospholipid synthesis had moderate up-regulation in expression and % relative mRNA abundance. The mRNA abundance and up-regulation in expression of ABCG2 during lactation was markedly high, suggesting a biological role of this gene in milk synthesis/secretion. Weak correlations were observed between both milk FA composition and desaturase indexes (i.e., apparent SCD activity) with mRNA expression pattern of genes measured.
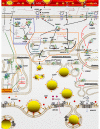
Conclusion: A network of genes participates in coordinating milk fat synthesis and secretion. Results challenge the proposal that SREBF1 is central for milk fat synthesis regulation and highlight a pivotal role for a concerted action among PPARG, PPARGC1A, and INSIG1. Expression of SCD, the most abundant gene measured, appears to be key during milk fat synthesis. The lack of correlation between gene expression and calculated desaturase indexes does not support their use to infer mRNA expression or enzyme activity (e.g., SCD). Longitudinal mRNA expression allowed development of transcriptional regulation networks and an updated model of milk fat synthesis regulation.
Figures


References
-
- Bauman DE, Mather IH, Wall RJ, Lock AL. Major advances associated with the biosynthesis of milk. J Dairy Sci. 2006;89:1235–1243. - PubMed
-
- Bauman DE, Davis CL. Biosynthesis of milk fat. In: Larson BL, Smith VR, editor. Lactation: a comprehensive treatise. Vol. 2. New York: Academic Press; 1974. pp. 31–75.
-
- Keenan TW, Mather IH. Intracellular Origin of Milk Fat Globules and the Nature of the Milk Fat Globule Membrane. In: Fox PF, McSweeney PLH, editor. Advanced Dairy Chemistry: Lipids. Vol. 2. New York, NY: Springer; 2006. pp. 137–171.
-
- Kay JK, Weber WJ, Moore CE, Bauman DE, Hansen LB, Chester-Jones H, Crooker BA, Baumgard LH. Effects of week of lactation and genetic selection for milk yield on milk fatty acid composition in Holstein cows. J Dairy Sci. 2005;88:3886–3893. - PubMed
-
- Garnsworthy PC, Masson LL, Lock AL, Mottram TT. Variation of milk citrate with stage of lactation and de novo fatty acid synthesis in dairy cows. J Dairy Sci. 2006;89:1604–1612. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

