Detailed kinetics and regulation of mammalian NAD-linked isocitrate dehydrogenase
- PMID: 18672100
- PMCID: PMC2584801
- DOI: 10.1016/j.bbapap.2008.07.001
Detailed kinetics and regulation of mammalian NAD-linked isocitrate dehydrogenase
Abstract
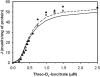
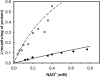
A mathematical model is presented to describe the catalytic mechanism of mammalian NAD-linked isocitrate dehydrogenase (NAD-IDH), a highly regulated enzyme in the tricarboxylic acid cycle, a crucial pathway in energy metabolism and biosynthesis. The mechanism accounts for allosteric regulation by magnesium-bound isocitrate and EGTA and calcium-bound ATP and ADP. The developed model is used to analyze kinetic data for the cardiac enzyme and to estimate kinetic parameter values. Since the kinetic mechanism is expressed in terms of chemical species (rather than biochemical reactants), the model explicitly accounts for the effects of biochemical state (ionic strength, pH, temperature, and metal cation concentration) on the kinetics. Because the substrate isocitrate competes with allosteric activators (ATP and ADP) and an inhibitor (EGTA) for metal ion cofactors (Ca(2+) and Mg(2+)), the observed kinetic relationships between reactants, activator and inhibitor concentrations, and catalytic flux are complex. Our analysis reveals that under physiological conditions, the ADP/ATP ratio plays a more significant role than Ca(2+) concentration in regulating the enzyme's activity. In addition, the enzyme is highly sensitive to Mg(2+) concentration in the physiological range, pointing to a potential regulatory role of [Mg(2+)] in mitochondrial energy metabolism.
Figures











References
-
- Hansford RG. Relation between mitochondrial calcium transport and control of energy metabolism. Rev Physiol Biochem Pharmacol. 1985;102:1–72. - PubMed
-
- Williamson JR, Cooper RH. Regulation of the citric acid cycle in mammalian systems. FEBS Lett. 1980;117(Suppl):K73–85. - PubMed
-
- Gabriel JL, Zervos PR, Plaut GW. Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria. Metabolism. 1986;35(7):661–667. - PubMed
-
- Plaut GW, Sung SC. Diphosphopyridine nucleotide isocitric dehydrogenase from animal tissues. J Biol Chem. 1954;207(1):305–314. - PubMed
-
- Goebell H, Klingenberg M. [Dpn-Specific Isocitrate Dehydrogenase of Mitochondria. I. Kinetic Properties, Occurrence and Function of Dpn-Specific Isocitrate Dehydrogenase.]. Biochem Z. 1964;340:441–464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

