In vitro dimerization of the bovine papillomavirus E5 protein transmembrane domain
- PMID: 18672907
- PMCID: PMC3711124
- DOI: 10.1021/bi8006252
In vitro dimerization of the bovine papillomavirus E5 protein transmembrane domain
Abstract
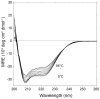
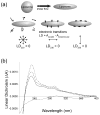
The E5 protein from bovine papillomavirus is a type II membrane protein and the product of the smallest known oncogene. E5 causes cell transformation by binding and activating the platelet-derived growth factor beta receptor (PDGFbetaR). In order to productively interact with the receptor, it is thought that E5 binds as a dimer. However, wild-type E5 and various mutants have also been shown to form trimers, tetramers, and even higher order oligomers. The residues in E5 that drive and stabilize a dimeric state are also still in question. At present, two different models for the E5 dimer exist in the literature, one symmetric and one asymmetric. There is universal agreement, however, that the transmembrane (TM) domain plays a vital role in stabilizing the functional oligomer; indeed, mutation of various TM domain residues can abolish E5 function. In order to better resolve the role of the E5 TM domain in function, we have undertaken the first quantitative in vitro characterization of the E5 TM domain in detergent micelles and liposomes. Circular and linear dichroism analyses verify that the TM domain adopts a stable alpha-helical structure and is able to partition efficiently across lipid bilayers. SDS-PAGE and analytical ultracentrifugation demonstrate for the first time that the TM domain of E5 forms a strong dimer with a standard state free energy of dissociation of 5.0 kcal mol (-1). We have used our new results to interpret existing models of E5 dimer formation and provide a direct link between TM helix interactions and E5 function.
Figures




References
-
- Drummond-Barbosa D, DiMaio D. Virocrine transformation. Biochem Biophys Acta. 1997;1332:M1–M17. - PubMed
-
- DiMaio D, Lai CC, Klein O. Virocrine transformation: the intersection between viral transforming proteins and cellular signal transduction pathways. Annual review of microbiology. 1998;52:397–421. - PubMed
-
- DiMaio D, Lai CC, Mattoon D. The platelet-derived growth factor beta receptor as a target of the bovine papillomavirus E5 protein. Cytokine Growth Factor Rev. 2000;11:283–293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

