Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae
- PMID: 18673560
- PMCID: PMC2535606
- DOI: 10.1186/1471-2164-9-370
Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae
Abstract
Background: The Zap1 transcription factor is a central player in the response of yeast to changes in zinc status. We previously used transcriptome profiling with DNA microarrays to identify 46 potential Zap1 target genes in the yeast genome. In this new study, we used complementary methods to identify additional Zap1 target genes.
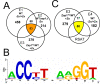
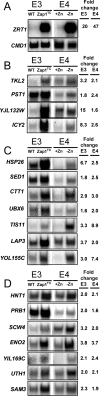
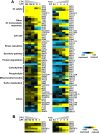
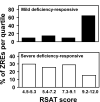
Results: With alternative growth conditions for the microarray experiments and a more sensitive motif identification algorithm, we identified 31 new potential targets of Zap1 activation. Moreover, an analysis of the response of Zap1 target genes to a range of zinc concentrations and to zinc withdrawal over time demonstrated that these genes respond differently to zinc deficiency. Some genes are induced under mild zinc deficiency and act as a first line of defense against this stress. First-line defense genes serve to maintain zinc homeostasis by increasing zinc uptake, and by mobilizing and conserving intracellular zinc pools. Other genes respond only to severe zinc limitation and act as a second line of defense. These second-line defense genes allow cells to adapt to conditions of zinc deficiency and include genes involved in maintaining secretory pathway and cell wall function, and stress responses.
Conclusion: We have identified several new targets of Zap1-mediated regulation. Furthermore, our results indicate that through the differential regulation of its target genes, Zap1 prioritizes mechanisms of zinc homeostasis and adaptive responses to zinc deficiency.
Figures
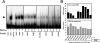

Similar articles
-
Zap1-dependent transcription from an alternative upstream promoter controls translation of RTC4 mRNA in zinc-deficient Saccharomyces cerevisiae.Mol Microbiol. 2017 Dec;106(5):678-689. doi: 10.1111/mmi.13851. Epub 2017 Oct 23. Mol Microbiol. 2017. PMID: 28963784 Free PMC article.
-
Roles of two activation domains in Zap1 in the response to zinc deficiency in Saccharomyces cerevisiae.J Biol Chem. 2011 Feb 25;286(8):6844-54. doi: 10.1074/jbc.M110.203927. Epub 2010 Dec 22. J Biol Chem. 2011. PMID: 21177862 Free PMC article.
-
Zap1 activation domain 1 and its role in controlling gene expression in response to cellular zinc status.Mol Microbiol. 2005 Aug;57(3):834-46. doi: 10.1111/j.1365-2958.2005.04734.x. Mol Microbiol. 2005. PMID: 16045625
-
Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae.J Biol Chem. 2009 Jul 10;284(28):18565-9. doi: 10.1074/jbc.R900014200. Epub 2009 Apr 10. J Biol Chem. 2009. PMID: 19363031 Free PMC article. Review.
-
Zinc sensing and regulation in yeast model systems.Arch Biochem Biophys. 2016 Dec 1;611:30-36. doi: 10.1016/j.abb.2016.02.031. Epub 2016 Mar 3. Arch Biochem Biophys. 2016. PMID: 26940262 Free PMC article. Review.
Cited by
-
Involvement of Sulfur in the Biosynthesis of Essential Metabolites in Pathogenic Fungi of Animals, Particularly Aspergillus spp.: Molecular and Therapeutic Implications.Front Microbiol. 2019 Dec 13;10:2859. doi: 10.3389/fmicb.2019.02859. eCollection 2019. Front Microbiol. 2019. PMID: 31921039 Free PMC article. Review.
-
Genomic Characterization of the Zinc Transcriptional Regulatory Element Reveals Potential Functional Roles of ZNF658.Biol Trace Elem Res. 2019 Dec;192(2):83-90. doi: 10.1007/s12011-019-1650-9. Epub 2019 Feb 7. Biol Trace Elem Res. 2019. PMID: 30734197 Free PMC article.
-
Effect of zinc deprivation on the lipid metabolism of budding yeast.Curr Genet. 2017 Dec;63(6):977-982. doi: 10.1007/s00294-017-0704-9. Epub 2017 May 12. Curr Genet. 2017. PMID: 28500379 Review.
-
Cytosolic superoxide dismutase (SOD1) is critical for tolerating the oxidative stress of zinc deficiency in yeast.PLoS One. 2009 Sep 16;4(9):e7061. doi: 10.1371/journal.pone.0007061. PLoS One. 2009. PMID: 19756144 Free PMC article.
-
Metals in fungal virulence.FEMS Microbiol Rev. 2018 Jan 1;42(1):fux050. doi: 10.1093/femsre/fux050. FEMS Microbiol Rev. 2018. PMID: 29069482 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

