Comparison of nonhomologous end joining and homologous recombination in human cells
- PMID: 18675941
- PMCID: PMC2695993
- DOI: 10.1016/j.dnarep.2008.06.018
Comparison of nonhomologous end joining and homologous recombination in human cells
Abstract
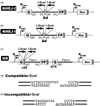
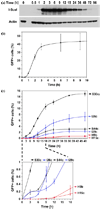
The two major pathways for repair of DNA double-strand breaks (DSBs) are homologous recombination (HR) and nonhomologous end joining (NHEJ). HR leads to accurate repair, while NHEJ is intrinsically mutagenic. To understand human somatic mutation it is essential to know the relationship between these pathways in human cells. Here we provide a comparison of the kinetics and relative contributions of HR and NHEJ in normal human cells. We used chromosomally integrated fluorescent reporter substrates for real-time in vivo monitoring of the NHEJ and HR. By examining multiple integrated clones we show that the efficiency of NHEJ and HR is strongly influenced by chromosomal location. Furthermore, we show that NHEJ of compatible ends (NHEJ-C) and NHEJ of incompatible ends (NHEJ-I) are fast processes, which can be completed in approximately 30 min, while HR is much slower and takes 7h or longer to complete. In actively cycling cells NHEJ-C is twice as efficient as NHEJ-I, and NHEJ-I is three times more efficient than HR. Our results suggest that NHEJ is a faster and more efficient DSB repair pathway than HR.
Conflict of interest statement
None.
Figures

References
-
- Haber JE. Partners and pathways repairing a double-strand break. Trends Genet. 2000;16:259–264. - PubMed
-
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. - PubMed
-
- Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 2001;477:131–153. - PubMed
-
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst.) 2006;5:1021–1029. - PubMed
-
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

