Calmodulin association with connexin32-derived peptides suggests trans-domain interaction in chemical gating of gap junction channels
- PMID: 18676375
- PMCID: PMC2555998
- DOI: 10.1074/jbc.M801434200
Calmodulin association with connexin32-derived peptides suggests trans-domain interaction in chemical gating of gap junction channels
Abstract
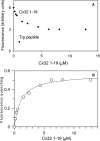
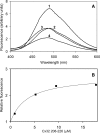
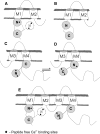
Calmodulin plays a key role in the chemical gating of gap junction channels. Two calmodulin-binding regions have previously been identified in connexin32 gap junction protein, one in the N-terminal and another in the C-terminal cytoplasmic tail of the molecule. The aim of this study was to better understand how calmodulin interacts with the connexin32-binding domains. Lobe-specific interactions of calmodulin with connexin32 peptides were studied by stopped flow kinetics, using Ca(2+) binding-deficient mutants. Peptides corresponding to the N-terminal tail (residues 1-22) of connexin32 engaged both the N- and C-terminal lobes (N- and C-lobes) of calmodulin, binding with higher affinity to the C-lobe of calmodulin (Ca(2+) dissociation rate constants k(3,4), 1.7+/-0.5 s(-1)) than to the N-lobe (k(1,2), 10.8+/-1.3 s(-1)). In contrast, peptides representing the C-terminal tail domain (residues 208-227) of connexin32 bound either the C- or the N-lobe but only one calmodulin lobe at a time (k(3,4), 2.6+/-0.1 s(-1) or k(1), 13.8+/-0.5 s(-1) and k(2), 1000 s(-1)). The calmodulin-binding domains of the N- and C-terminal tails of connexin32 were best defined as residues 1-21 and 216-227, respectively. Our data, showing separate functions of the N- and C-lobes of calmodulin in the interactions with connexin32, suggest trans-domain or trans-subunit bridging by calmodulin as a possible mechanism of gap junction gating.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

