The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation
- PMID: 18676449
- PMCID: PMC2528179
- DOI: 10.1093/nar/gkn484
The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation
Abstract
The mammalian architectural HMGB-Box transcription factor UBF is ubiquitously expressed in two variant forms as the result of a differential splicing event, that in the UBF2 deletes 37 amino acid from the second of six HMGB-boxes. Several attempts to define a function for this shorter UBF2 protein have been less than satisfactory. However, since all mammals appear to display similar levels of the longer and shorter UBF variants, it is unlikely that UBF2 is simply nonfunctional. Previously we showed that phosphorylation of UBF by the MAP-kinase ERK regulates chromatin folding and transcription elongation, explaining the rapid response of the ribosomal RNA genes to growth factors. Here we have investigated the roles the UBF variants play in the response of these genes to ERK activity. We demonstrate that the variant HMGB-box 2 of UBF2 has lost the ability to bind bent DNA and hence to induce chromatin folding. As a result it is significantly less effective than UBF1 at arresting RNAPI elongation but at the same time is more responsive to ERK phosphorylation. Thus, UBF2 functionally simulates a hemi-phosphorylated UBF whose expression may provide a means by which to tune the response of the ribosomal RNA genes to growth factor stimulation.
Figures




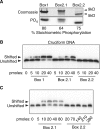


References
-
- Jantzen H-M, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

