Phage-associated mutator phenotype in group A streptococcus
- PMID: 18676670
- PMCID: PMC2565987
- DOI: 10.1128/JB.01569-07
Phage-associated mutator phenotype in group A streptococcus
Abstract
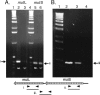
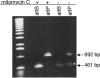
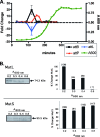
Defects in DNA mismatch repair (MMR) occur frequently in natural populations of pathogenic and commensal bacteria, resulting in a mutator phenotype. We identified a unique genetic element in Streptococcus pyogenes strain SF370 that controls MMR via a dynamic process of prophage excision and reintegration in response to growth. In S. pyogenes, mutS and mutL are organized on a polycistronic mRNA under control of a common promoter. Prophage SF370.4 is integrated between the two genes, blocking expression of the downstream gene (mutL) and resulting in a mutator phenotype. However, in rapidly growing cells the prophage excises and replicates as an episome, allowing mutL to be expressed. Excision of prophage SF370.4 and expression of MutL mRNA occur simultaneously during early logarithmic growth when cell densities are low; this brief window of MutL gene expression ends as the cell density increases. However, detectable amounts of MutL protein remain in the cell until the onset of stationary phase. Thus, MMR in S. pyogenes SF370 is functional in exponentially growing cells but defective when resources are limiting. The presence of a prophage integrated into the 5' end of mutL correlates with a mutator phenotype (10(-7) to 10(-8) mutation/generation, an approximately a 100-fold increase in the rate of spontaneous mutation compared with prophage-free strains [10(-9) to 10(-10) mutation/generation]). Such genetic elements may be common in S. pyogenes since 6 of 13 completed genomes have related prophages, and a survey of 100 strains found that about 20% of them are positive for phages occupying the SF370.4 attP site. The dynamic control of a major DNA repair system by a bacteriophage is a novel method for achieving the mutator phenotype and may allow the organism to respond rapidly to a changing environment while minimizing the risks associated with long-term hypermutability.
Figures




Similar articles
-
The Bacteriophages of Streptococcus pyogenes.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0059-2018. doi: 10.1128/microbiolspec.GPP3-0059-2018. Microbiol Spectr. 2019. PMID: 31111820 Free PMC article. Review.
-
Genome analysis of an inducible prophage and prophage remnants integrated in the Streptococcus pyogenes strain SF370.Virology. 2002 Oct 25;302(2):245-58. doi: 10.1006/viro.2002.1570. Virology. 2002. PMID: 12441069
-
Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic Streptococci: evolutionary implications for prophage-host interactions.Virology. 2001 Sep 30;288(2):325-41. doi: 10.1006/viro.2001.1085. Virology. 2001. PMID: 11601904
-
Phage-Like Streptococcus pyogenes Chromosomal Islands (SpyCI) and Mutator Phenotypes: Control by Growth State and Rescue by a SpyCI-Encoded Promoter.Front Microbiol. 2012 Aug 30;3:317. doi: 10.3389/fmicb.2012.00317. eCollection 2012. Front Microbiol. 2012. PMID: 22969756 Free PMC article.
-
Chromosomal islands of Streptococcus pyogenes and related streptococci: molecular switches for survival and virulence.Front Cell Infect Microbiol. 2014 Aug 12;4:109. doi: 10.3389/fcimb.2014.00109. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25161960 Free PMC article. Review.
Cited by
-
The Bacteriophages of Streptococcus pyogenes.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0059-2018. doi: 10.1128/microbiolspec.GPP3-0059-2018. Microbiol Spectr. 2019. PMID: 31111820 Free PMC article. Review.
-
The structural characterization of a prophage-encoded extracellular DNase from Streptococcus pyogenes.Nucleic Acids Res. 2012 Jan;40(2):928-38. doi: 10.1093/nar/gkr789. Epub 2011 Sep 24. Nucleic Acids Res. 2012. PMID: 21948797 Free PMC article.
-
A novel conjugative plasmid from Enterococcus faecalis E99 enhances resistance to ultraviolet radiation.Plasmid. 2010 Jul;64(1):18-25. doi: 10.1016/j.plasmid.2010.03.001. Epub 2010 Mar 20. Plasmid. 2010. PMID: 20307569 Free PMC article.
-
Bacterial genome instability.Microbiol Mol Biol Rev. 2014 Mar;78(1):1-39. doi: 10.1128/MMBR.00035-13. Microbiol Mol Biol Rev. 2014. PMID: 24600039 Free PMC article. Review.
-
Long-term survival of Streptococcus pyogenes in rich media is pH-dependent.Microbiology (Reading). 2012 Jun;158(Pt 6):1428-1436. doi: 10.1099/mic.0.054478-0. Epub 2012 Feb 23. Microbiology (Reading). 2012. PMID: 22361943 Free PMC article.
References
-
- Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggard-Ljungquist, R. H. Hoess, M. L. Kahn, B. Kalionis, S. V. L. Narayana, L. S. Pierson III, N. Sternberg, and J. M. Leong. 1986. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5433-440. - PMC - PubMed
-
- Banks, D. J., S. F. Porcella, K. D. Barbian, S. B. Beres, L. E. Philips, J. M. Voyich, F. R. DeLeo, J. M. Martin, G. A. Somerville, and J. M. Musser. 2004. Progress toward characterization of the group A streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190727-738. - PubMed
-
- Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 9910078-10083. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

