Monitoring abundance and expression of "Dehalococcoides" species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column
- PMID: 18676701
- PMCID: PMC2547056
- DOI: 10.1128/AEM.00926-08
Monitoring abundance and expression of "Dehalococcoides" species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column
Abstract
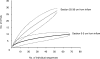
We investigated the distribution and activity of chloroethene-degrading microorganisms and associated functional genes during reductive dehalogenation of tetrachloroethene to ethene in a laboratory continuous-flow column. Using real-time PCR, we quantified "Dehalococcoides" species 16S rRNA and chloroethene-reductive dehalogenase (RDase) genes (pceA, tceA, vcrA, and bvcA) in nucleic acid extracts from different sections of the column. Dehalococcoides 16S rRNA gene copies were highest at the inflow port [(3.6 +/- 0.6) x 10(6) (mean +/- standard deviation) per gram soil] where the electron donor and acceptor were introduced into the column. The highest transcript numbers for tceA, vcrA, and bvcA were detected 5 to 10 cm from the column inflow. bvcA was the most highly expressed of all RDase genes and the only vinyl chloride reductase-encoding transcript detectable close to the column outflow. Interestingly, no expression of pceA was detected in the column, despite the presence of the genes in the microbial community throughout the column. By comparing the 16S rRNA gene copy numbers to the sum of all four RDase genes, we found that 50% of the Dehalococcoides population in the first part of the column did not contain either one of the known chloroethene RDase genes. Analysis of 16S rRNA gene clone libraries from both ends of the flow column revealed a microbial community dominated by members of Firmicutes and Actinobacteria. Higher clone sequence diversity was observed near the column outflow. The results presented have implications for our understanding of the ecophysiology of reductively dehalogenating Dehalococcoides spp. and their role in bioremediation of chloroethenes.
Figures
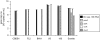


References
-
- Adrian, L., U. Szewzyk, J. Wecke, and H. Gorisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. - PubMed
-
- Azizian, M. F., S. Behrens, A. Sabalowsky, M. E. Dolan, A. M. Spormann, and L. Semprini. 2008. Continuous-flow column study of reductive dehalogenation of PCE upon bioaugmentation with the Evanite enrichment culture. J. Contam. Hydrol. 100:11-21. - PubMed
-
- Duhamel, M., and E. A. Edwards. 2006. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol. Ecol. 58:538-549. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

