Preferential colonization of Solanum tuberosum L. roots by the fungus Glomus intraradices in arable soil of a potato farming area
- PMID: 18676711
- PMCID: PMC2547043
- DOI: 10.1128/AEM.00719-08
Preferential colonization of Solanum tuberosum L. roots by the fungus Glomus intraradices in arable soil of a potato farming area
Abstract
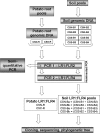
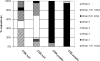
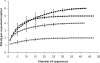
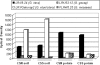
The symbiosis between plant roots and arbuscular mycorrhizal (AM) fungi has been shown to affect both the diversity and productivity of agricultural communities. In this study, we characterized the AM fungal communities of Solanum tuberosum L. (potato) roots and of the bulk soil in two nearby areas of northern Italy, in order to verify if land use practices had selected any particular AM fungus with specificity to potato plants. The AM fungal large-subunit (LSU) rRNA genes were subjected to nested PCR, cloning, sequencing, and phylogenetic analyses. One hundred eighty-three LSU rRNA sequences were analyzed, and eight monophyletic ribotypes, belonging to Glomus groups A and B, were identified. AM fungal communities differed between bulk soil and potato roots, as one AM fungal ribotype, corresponding to Glomus intraradices, was much more frequent in potato roots than in soils (accounting for more than 90% of sequences from potato samples and less than 10% of sequences from soil samples). A semiquantitative heminested PCR with specific primers was used to confirm and quantify the AM fungal abundance observed by cloning. Overall results concerning the biodiversity of AM fungal communities in roots and in bulk soils from the two studied areas suggested that potato roots were preferentially colonized by one AM fungal species, G. intraradices.
Figures

References
-
- Bever, J. D. 2002. Host-specificity of AM fungal population growth rates can generate feedback on plant growth. Plant Soil 244:281-290.
-
- Bever, J. D., A. Pringle, and P. A. Schultz. 2002. Dynamics within the plant-arbuscular mycorrhizal fungal mutualism: testing the nature of community feedback, p. 267-292. In M. G. A. van der Heijden and I. E. Sanders (ed.), Mycorrhizal ecology. Springer-Verlag, Berlin, Germany.
-
- Bharadwaj, D. P., P. Lundquist, and S. Alströma. 2007. Impact of plant species grown as monocultures on sporulation and root colonization by native arbuscular mycorrhizal fungi in potato. Appl. Soil Ecol. 35:213-225.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

