An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer
- PMID: 18676830
- PMCID: PMC2680495
- DOI: 10.1158/0008-5472.CAN-07-6854
An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer
Abstract
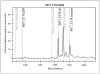
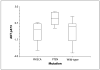
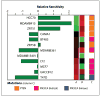
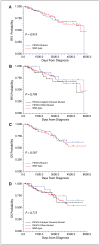
Phosphatidylinositol 3-kinase (PI3K)/AKT pathway aberrations are common in cancer. By applying mass spectroscopy-based sequencing and reverse-phase protein arrays to 547 human breast cancers and 41 cell lines, we determined the subtype specificity and signaling effects of PIK3CA, AKT, and PTEN mutations and the effects of PIK3CA mutations on responsiveness to PI3K inhibition in vitro and on outcome after adjuvant tamoxifen. PIK3CA mutations were more common in hormone receptor-positive (34.5%) and HER2-positive (22.7%) than in basal-like tumors (8.3%). AKT1 (1.4%) and PTEN (2.3%) mutations were restricted to hormone receptor-positive cancers. Unlike AKT1 mutations that were absent from cell lines, PIK3CA (39%) and PTEN (20%) mutations were more common in cell lines than tumors, suggesting a selection for these but not AKT1 mutations during adaptation to culture. PIK3CA mutations did not have a significant effect on outcome after adjuvant tamoxifen therapy in 157 hormone receptor-positive breast cancer patients. PIK3CA mutations, in comparison with PTEN loss and AKT1 mutations, were associated with significantly less and inconsistent activation of AKT and of downstream PI3K/AKT signaling in tumors and cell lines. PTEN loss and PIK3CA mutation were frequently concordant, suggesting different contributions to pathophysiology. PTEN loss rendered cells significantly more sensitive to growth inhibition by the PI3K inhibitor LY294002 than did PIK3CA mutations. Thus, PI3K pathway aberrations likely play a distinct role in the pathogenesis of different breast cancer subtypes. The specific aberration present may have implications for the selection of PI3K-targeted therapies in hormone receptor-positive breast cancer.
Conflict of interest statement
J.W. Gray: commercial research grants, GlaxoSmithKline, Cellgate, Affymetrix, and Cell Biosciences; consultant, Agendia, Cepheid, and Bristol-Myers Squibb. G.B. Mills: scientific/advisory committee member, Abbott Laboratories, Ambit Biosciences Corp., Lpath Therapeutics Inc., and Texas Institute for Genomic Medicine; consultant, GlaxoSmithKline, Semafore Pharmaceuticals Inc., and TAU Therapeutics; stock options, GLT, Inc.; royalty income, Upstate Biotechnology. The other authors disclosed no potential conflicts of interest.
Figures

References
-
- Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. - PubMed
-
- Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. - PubMed
-
- Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30CA16672/CA/NCI NIH HHS/United States
- R01 CA109311/CA/NCI NIH HHS/United States
- P01CA099031/CA/NCI NIH HHS/United States
- U54 CA 112970/CA/NCI NIH HHS/United States
- 1K23CA121994-01/CA/NCI NIH HHS/United States
- P50 CA058207/CA/NCI NIH HHS/United States
- U54 CA112970/CA/NCI NIH HHS/United States
- P50 CA083639/CA/NCI NIH HHS/United States
- P01 CA099031/CA/NCI NIH HHS/United States
- P50CA083639/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- K23 CA121994/CA/NCI NIH HHS/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- 1R21CA120248-01/CA/NCI NIH HHS/United States
- P50 CA 58207/CA/NCI NIH HHS/United States
- R21 CA120248/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

