Par-4 binds to topoisomerase 1 and attenuates its DNA relaxation activity
- PMID: 18676842
- PMCID: PMC2562756
- DOI: 10.1158/0008-5472.CAN-08-0831
Par-4 binds to topoisomerase 1 and attenuates its DNA relaxation activity
Abstract
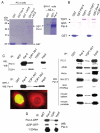
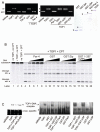
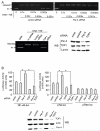
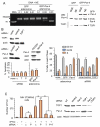
The regulation of DNA relaxation by topoisomerase 1 (TOP1) is essential for DNA replication, transcription, and recombination events. TOP1 activity is elevated in cancer cells, yet the regulatory mechanism restraining its activity is not understood. We present evidence that the tumor suppressor protein prostate apoptosis response-4 (Par-4) directly binds to TOP1 and attenuates its DNA relaxation activity. Unlike camptothecin, which binds at the TOP1-DNA interface to form cleavage complexes, Par-4 interacts with TOP1 via its leucine zipper domain and sequesters TOP1 from the DNA. Par-4 knockdown by RNA interference enhances DNA relaxation and gene transcription activities and promotes cellular transformation in a TOP1-dependent manner. Conversely, attenuation of TOP1 activity either by RNA interference or Par-4 overexpression impedes DNA relaxation, cell cycle progression, and gene transcription activities and inhibits transformation. Collectively, our findings suggest that Par-4 serves as an intracellular repressor of TOP1 catalytic activity and regulates DNA topology to suppress cellular transformation.
Figures
References
-
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. - PubMed
-
- Koster DA, Palle K, Bot ES, et al. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–7. - PubMed
-
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. - PubMed
-
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

