Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice
- PMID: 18676878
- PMCID: PMC2553609
- DOI: 10.1105/tpc.107.057257
Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice
Abstract
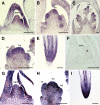
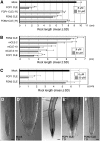
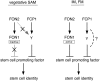
Postembryonic development in plants depends on the activity of the shoot apical meristem (SAM) and root apical meristem (RAM). In Arabidopsis thaliana, CLAVATA signaling negatively regulates the size of the stem cell population in the SAM by repressing WUSCHEL. In other plants, however, studies of factors involved in stem cell maintenance are insufficient. Here, we report that two proteins closely related to CLAVATA3, FLORAL ORGAN NUMBER2 (FON2) and FON2-LIKE CLE PROTEIN1 (FCP1/Os CLE402), have functionally diversified to regulate the different types of meristem in rice (Oryza sativa). Unlike FON2, which regulates the maintenance of flower and inflorescence meristems, FCP1 appears to regulate the maintenance of the vegetative SAM and RAM. Constitutive expression of FCP1 results in consumption of the SAM in the vegetative phase, and application of an FCP1 CLE peptide in vitro disturbs root development by misspecification of cell fates in the RAM. FON1, a putative receptor of FON2, is likely to be unnecessary for these FCP1 functions. Furthermore, we identify a key amino acid residue that discriminates between the actions of FCP1 and FON2. Our results suggest that, although the basic framework of meristem maintenance is conserved in the angiosperms, the functions of the individual factors have diversified during evolution.
Figures



Similar articles
-
Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene.Plant Cell Physiol. 2006 Dec;47(12):1591-602. doi: 10.1093/pcp/pcl025. Epub 2006 Oct 20. Plant Cell Physiol. 2006. PMID: 17056620
-
WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice.Plant Cell. 2013 Jan;25(1):229-41. doi: 10.1105/tpc.112.103432. Epub 2013 Jan 31. Plant Cell. 2013. PMID: 23371950 Free PMC article.
-
FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice.PLoS Genet. 2009 Oct;5(10):e1000693. doi: 10.1371/journal.pgen.1000693. Epub 2009 Oct 16. PLoS Genet. 2009. PMID: 19834537 Free PMC article.
-
Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem.J Plant Res. 2009 Jan;122(1):31-9. doi: 10.1007/s10265-008-0207-3. Epub 2008 Dec 23. J Plant Res. 2009. PMID: 19104754 Review.
-
CLE peptides: critical regulators for stem cell maintenance in plants.Planta. 2021 Nov 29;255(1):5. doi: 10.1007/s00425-021-03791-1. Planta. 2021. PMID: 34841457 Review.
Cited by
-
Evolution of meristem zonation by CLE gene duplication in land plants.Nat Plants. 2022 Jul;8(7):735-740. doi: 10.1038/s41477-022-01199-7. Epub 2022 Jul 19. Nat Plants. 2022. PMID: 35854003 Review.
-
Genome-wide characterization, expression and functional analysis of CLV3/ESR gene family in tomato.BMC Genomics. 2014 Sep 30;15(1):827. doi: 10.1186/1471-2164-15-827. BMC Genomics. 2014. PMID: 25266499 Free PMC article.
-
Novel insights into maize (Zea mays) development and organogenesis for agricultural optimization.Planta. 2023 Apr 9;257(5):94. doi: 10.1007/s00425-023-04126-y. Planta. 2023. PMID: 37031436 Review.
-
The Rice Receptor-Like Kinases DWARF AND RUNTISH SPIKELET1 and 2 Repress Cell Death and Affect Sugar Utilization during Reproductive Development.Plant Cell. 2017 Jan;29(1):70-89. doi: 10.1105/tpc.16.00218. Epub 2017 Jan 12. Plant Cell. 2017. PMID: 28082384 Free PMC article.
-
CLE peptide signaling during plant development.Protoplasma. 2010 Apr;240(1-4):33-43. doi: 10.1007/s00709-009-0095-y. Epub 2009 Dec 17. Protoplasma. 2010. PMID: 20016993 Free PMC article. Review.
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Bommert, P., Lunde, C., Nardmann, J., Vollbrecht, E., Running, M., Jackson, D., Hake, S., and Werr, W. (2005. a). thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132 1235–1245. - PubMed
-
- Bommert, P., Satoh-Nagasawa, N., Jackson, D., and Hirano, H.-Y. (2005. b). Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46 69–78. - PubMed
-
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619. - PubMed
-
- Casamitjana-Martinez, E., Hofhuis, H.F., Xu, J., Liu, C.-M., Heidstra, R., and Scheres, B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13 1435–1441. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

