Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells
- PMID: 18678384
- PMCID: PMC7103401
- DOI: 10.1016/j.virol.2008.06.038
Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells
Abstract
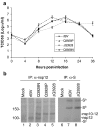
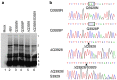
Coronavirus 3C-like proteinase (3CLpro) plays important roles in viral life cycle through extensive processing of the polyproteins 1a and 1ab into 12 mature, non-structural proteins (nsp5-nsp16). Structural and biochemical studies have revealed that all confirmed 3CLpro cleavage sites have a conserved Gln residue at the P1 position, which is thought to be absolutely required for efficient cleavage. Recent studies on murine hepatitis virus (MHV) showed that processing of the 1a polyprotein at the position between nsp10-nsp11 is essential for viral replication. In this report, we investigated the requirement of processing at the equivalent position for replication of avian coronavirus infectious bronchitis virus (IBV), using an infectious cloning system. The results showed that mutation of the P1 Gln to Pro or deletion of the Gln residue in the nsp10-nsp11/12 site completely abolished the 3CLpro-mediated processing, but allowed production of infectious recombinant viruses with variable degrees of growth defect, suggesting that cleavage at the nsp10-nsp11/12 site of IBV is dispensable for viral replication in cultured cells. This study would pave a way for potential vaccine development by generation of attenuated IBV from field isolates through manipulation of the nsp10-nsp11/12 cleavage site. Similar approaches would be also applicable to other human and animal coronaviruses.
Figures

References
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
-
- Boursnell M.E.G., Brown T.D.K., Foulds I.J., Green P.F., Tomely F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

