Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H
- PMID: 18678872
- PMCID: PMC2662066
- DOI: 10.1074/jbc.M803092200
Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H
Abstract
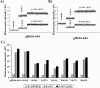
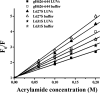
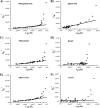
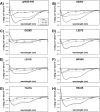
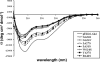
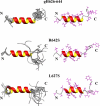
Glycoprotein H (gH) of herpes simplex virus type I (HSV-1) is involved in the complex mechanism of membrane fusion of the viral envelope with the host cell. Membrane interacting regions and potential fusion peptides have been identified in HSV-1 gH as well as glycoprotein B (gB). Because of the complex fusion mechanism of HSV-1, which requires four viral glycoproteins, and because there are only structural data for gB and glycoprotein D, many questions regarding the mechanism by which HSV-1 fuses its envelope with the host cell membrane remain unresolved. Previous studies have shown that peptides derived from certain regions of gH have the potential to interact with membranes, and based on these findings we have generated a set of peptides containing mutations in one of these domains, gH-(626-644), to investigate further the functional role of this region. Using a combination of biochemical, spectroscopic, and nuclear magnetic resonance techniques, we showed that the alpha-helical nature of this stretch of amino acids in gH is important for membrane interaction and that the aromatic residues, tryptophan and tyrosine, are critical for induction of fusion.
Figures



Similar articles
-
Role of membranotropic sequences from herpes simplex virus type I glycoproteins B and H in the fusion process.Biochim Biophys Acta. 2010 Mar;1798(3):579-91. doi: 10.1016/j.bbamem.2010.01.006. Epub 2010 Jan 18. Biochim Biophys Acta. 2010. PMID: 20085747
-
The presence of a single N-terminal histidine residue enhances the fusogenic properties of a Membranotropic peptide derived from herpes simplex virus type 1 glycoprotein H.J Biol Chem. 2010 May 28;285(22):17123-36. doi: 10.1074/jbc.M110.114819. Epub 2010 Mar 26. J Biol Chem. 2010. PMID: 20348105 Free PMC article.
-
Functional Characterization of Glycoprotein H Chimeras Composed of Conserved Domains of the Pseudorabies Virus and Herpes Simplex Virus 1 Homologs.J Virol. 2015 Oct 21;90(1):421-32. doi: 10.1128/JVI.01985-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491153 Free PMC article.
-
The role of herpes simplex virus glycoproteins in the virus replication cycle.Acta Virol. 1998 Apr;42(2):103-18. Acta Virol. 1998. PMID: 9770079 Review.
-
The role of glycoprotein H in herpesvirus membrane fusion.Protein Pept Lett. 2009;16(7):760-5. doi: 10.2174/092986609788681850. Protein Pept Lett. 2009. PMID: 19601905 Review.
Cited by
-
Interaction domain of glycoproteins gB and gH of Marek's disease virus and identification of an antiviral peptide with dual functions.PLoS One. 2013;8(2):e54761. doi: 10.1371/journal.pone.0054761. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405092 Free PMC article.
-
Exploitation of viral properties for intracellular delivery.J Pept Sci. 2014 Jul;20(7):468-78. doi: 10.1002/psc.2649. Epub 2014 May 30. J Pept Sci. 2014. PMID: 24889153 Free PMC article. Review.
-
gH625: a milestone in understanding the many roles of membranotropic peptides.Biochim Biophys Acta. 2015 Jan;1848(1 Pt A):16-25. doi: 10.1016/j.bbamem.2014.10.006. Epub 2014 Oct 12. Biochim Biophys Acta. 2015. PMID: 25305339 Free PMC article. Review.
-
Dissociation of HSV gL from gH by αvβ6- or αvβ8-integrin promotes gH activation and virus entry.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):E3901-10. doi: 10.1073/pnas.1506846112. Epub 2015 Jul 8. Proc Natl Acad Sci U S A. 2015. PMID: 26157134 Free PMC article.
-
gH625 is a viral derived peptide for effective delivery of intrinsically disordered proteins.Int J Nanomedicine. 2013;8:2555-65. doi: 10.2147/IJN.S44186. Epub 2013 Jul 22. Int J Nanomedicine. 2013. PMID: 23901273 Free PMC article.
References
-
- Montgomery, R. I., Warner, M. S., Lum, B. J., and Spear, P. G. (1996) Cell 87 427-436 - PubMed
-
- Geraghty, R. J., Krummenacher, C., Cohen, G. H., Eisenberg, R. J., and Spear, P. G. (1998) Science 280 1618-1620 - PubMed
-
- Warner, M. S., Geraghty, R. J., Martinez, W. M., Montgomery, R. I., Whitbeck, J. C., Xu, R., Eisenberg, R. J., Cohen, G. H., and Spear, P. G. (1998) Virology 246 179-189 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

