Dynamic energy landscape view of coupled binding and protein conformational change: induced-fit versus population-shift mechanisms
- PMID: 18678900
- PMCID: PMC2516237
- DOI: 10.1073/pnas.0802524105
Dynamic energy landscape view of coupled binding and protein conformational change: induced-fit versus population-shift mechanisms
Abstract
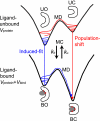
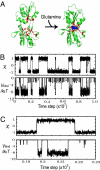
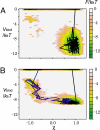
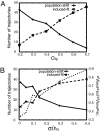
Allostery, the coupling between ligand binding and protein conformational change, is the heart of biological network and it has often been explained by two representative models, the induced-fit and the population-shift models. Here, we clarified for what systems one model fits better than the other by performing molecular simulations of coupled binding and conformational change. Based on the dynamic energy landscape view, we developed an implicit ligand-binding model combined with the double-basin Hamiltonian that describes conformational change. From model simulations performed for a broad range of parameters, we uncovered that each of the two models has its own range of applicability, stronger and longer-ranged interaction between ligand and protein favors the induced-fit model, and weaker and shorter-ranged interaction leads to the population-shift model. We further postulate that the protein binding to small ligand tends to proceed via the population-shift model, whereas the protein docking to macromolecules such as DNA tends to fit the induced-fit model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Qi G, Lee R, Hayward S. A comprehensive and non-redundant database of protein domain movements. Bioinformatics. 2005;21:2832–2838. - PubMed
-
- Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. - PubMed
-
- Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

