Ion permeation through a Cl--selective channel designed from a CLC Cl-/H+ exchanger
- PMID: 18678918
- PMCID: PMC2516207
- DOI: 10.1073/pnas.0804503105
Ion permeation through a Cl--selective channel designed from a CLC Cl-/H+ exchanger
Abstract
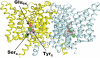
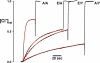
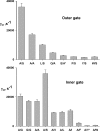
The CLC family of Cl(-)-transporting proteins includes both Cl(-) channels and Cl(-)/H(+) exchange transporters. CLC-ec1, a structurally known bacterial homolog of the transporter subclass, exchanges two Cl(-) ions per proton with strict, obligatory stoichiometry. Point mutations at two residues, Glu(148) and Tyr(445), are known to impair H(+) movement while preserving Cl(-) transport. In the x-ray crystal structure of CLC-ec1, these residues form putative "gates" flanking an ion-binding region. In mutants with both of the gate-forming side chains reduced in size, H(+) transport is abolished, and unitary Cl(-) transport rates are greatly increased, well above values expected for transporter mechanisms. Cl(-) transport rates increase as side-chain volume at these positions is decreased. The crystal structure of a doubly ungated mutant shows a narrow conduit traversing the entire protein transmembrane width. These characteristics suggest that Cl(-) flux through uncoupled, ungated CLC-ec1 occurs via a channel-like electrodiffusion mechanism rather than an alternating-exposure conformational cycle that has been rendered proton-independent by the gate mutations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

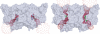
Similar articles
-
Intracellular proton-transfer mutants in a CLC Cl-/H+ exchanger.J Gen Physiol. 2009 Feb;133(2):131-8. doi: 10.1085/jgp.200810112. Epub 2009 Jan 12. J Gen Physiol. 2009. PMID: 19139174 Free PMC article.
-
A CLC-ec1 mutant reveals global conformational change and suggests a unifying mechanism for the CLC Cl-/H+ transport cycle.Elife. 2020 Apr 20;9:e53479. doi: 10.7554/eLife.53479. Elife. 2020. PMID: 32310757 Free PMC article.
-
Probing the conformation of a conserved glutamic acid within the Cl- pathway of a CLC H+/Cl- exchanger.J Gen Physiol. 2017 Apr 3;149(4):523-529. doi: 10.1085/jgp.201611682. Epub 2017 Feb 28. J Gen Physiol. 2017. PMID: 28246117 Free PMC article.
-
Structural insights into chloride and proton-mediated gating of CLC chloride channels.Biochemistry. 2004 Feb 10;43(5):1135-44. doi: 10.1021/bi0359776. Biochemistry. 2004. PMID: 14756549 Review.
-
CLC channels and transporters: proteins with borderline personalities.Biochim Biophys Acta. 2010 Aug;1798(8):1457-64. doi: 10.1016/j.bbamem.2010.02.022. Epub 2010 Feb 24. Biochim Biophys Acta. 2010. PMID: 20188062 Free PMC article. Review.
Cited by
-
Evolution of the genetic code by incorporation of amino acids that improved or changed protein function.J Mol Evol. 2013 Oct;77(4):134-58. doi: 10.1007/s00239-013-9567-y. Epub 2013 Jun 7. J Mol Evol. 2013. PMID: 23743924
-
Molecular mechanism of proton transport in CLC Cl-/H+ exchange transporters.Proc Natl Acad Sci U S A. 2012 Jul 17;109(29):11699-704. doi: 10.1073/pnas.1205764109. Epub 2012 Jul 2. Proc Natl Acad Sci U S A. 2012. PMID: 22753511 Free PMC article.
-
Voltage-dependent and -independent titration of specific residues accounts for complex gating of a ClC chloride channel by extracellular protons.J Physiol. 2009 Apr 1;587(Pt 7):1387-400. doi: 10.1113/jphysiol.2008.167353. Epub 2009 Jan 19. J Physiol. 2009. PMID: 19153159 Free PMC article.
-
Chloride channels: often enigmatic, rarely predictable.Annu Rev Physiol. 2010;72:95-121. doi: 10.1146/annurev-physiol-021909-135811. Annu Rev Physiol. 2010. PMID: 19827947 Free PMC article. Review.
-
Structure of a CLC chloride ion channel by cryo-electron microscopy.Nature. 2017 Jan 26;541(7638):500-505. doi: 10.1038/nature20812. Epub 2016 Dec 21. Nature. 2017. PMID: 28002411 Free PMC article.
References
-
- Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer; 2001.
-
- Gennis RB. Biomembranes: Molecular Structure and Function. New York: Springer; 1989.
-
- Gouaux E, MacKinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. - PubMed
-
- Matulef K, Maduke M. The CLC “chloride channel” family: Revelations from prokaryotes. Mol Membr Biol. 2007;24:342–350. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

