The multiple lives of NMD factors: balancing roles in gene and genome regulation
- PMID: 18679436
- PMCID: PMC3711694
- DOI: 10.1038/nrg2402
The multiple lives of NMD factors: balancing roles in gene and genome regulation
Abstract
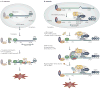
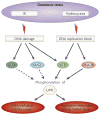
Nonsense-mediated mRNA decay (NMD) largely functions to ensure the quality of gene expression. However, NMD is also crucial to regulating appropriate expression levels for certain genes and for maintaining genome stability. Furthermore, just as NMD serves cells in multiple ways, so do its constituent proteins. Recent studies have clarified that UPF and SMG proteins, which were originally discovered to function in NMD, also have roles in other pathways, including specialized pathways of mRNA decay, DNA synthesis and cell-cycle progression, and the maintenance of telomeres. These findings suggest a delicate balance of metabolic events - some not obviously related to NMD - that can be influenced by the cellular abundance, location and activity of NMD factors and their binding partners.
Figures


References
-
- Behm-Ansmant I, et al. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. - PubMed
-
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. - PubMed
-
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. A comprehensive review of translation-dependent mRNA surveillance mechanisms, with a focus on NMD. - PubMed
-
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nature Genet. 2004;36:801–808. - PubMed
-
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nature Rev Mol Cell Biol. 2006;7:415–425. A comprehensive overview of NMD in S. cerevisiae. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

