Enhancement of p53 expression in keratinocytes by the bioflavonoid apigenin is associated with RNA-binding protein HuR
- PMID: 18680106
- PMCID: PMC2631086
- DOI: 10.1002/mc.20460
Enhancement of p53 expression in keratinocytes by the bioflavonoid apigenin is associated with RNA-binding protein HuR
Abstract
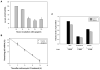
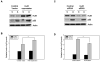
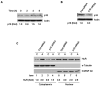
We have reported previously that apigenin, a naturally occurring nonmutagenic flavonoid, increased wild-type p53 protein expression in the mouse keratinocyte 308 cell line by a mechanism involving p53 protein stabilization. Here we further demonstrated that the increase in p53 protein level induced by apigenin treatment of 308 keratinoyctes was not the result of enhanced transcription, mRNA stabilization or cytoplasmic export of p53 mRNA. Instead, biosynthetic labeling showed that apigenin increased nascent p53 protein synthesis by enhancing p53 translation. The AU-rich element (ARE) within the 3'-untranslated region (UTR) of p53 mRNA was found to be responsible for apigenin's ability to increase p53 translation, as demonstrated in studies wherein the 3'-UTR of p53 mRNA containing the ARE was fused downstream of a luciferase reporter gene. Furthermore, apigenin treatment increased the level of association of the RNA binding protein HuR with endogenous p53 mRNA. Apigenin treatment also augmented HuR translocation into the cytoplasm. Overexpression of HuR enhanced apigenin-induced p53 protein expression in 308 keratinocytes, whereas siRNA-mediated HuR reduction suppressed apigenin-induced p53 protein expression and de novo translation of p53. Moreover, apigenin treatment of cells induced p16 protein expression, which in turn was correlated with cytoplasmic localization of HuR induced by apigenin. Overall, these findings indicate that, in addition to modulating p53 protein stability, one of the mechanisms by which apigenin induces p53 protein expression is enhancement of translation through the RNA binding protein HuR.
Copyright 2008 Wiley-Liss, Inc.
Figures







References
-
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. - PubMed
-
- Ryan KM, Phillips AC, Vousden KH. Regulation and function of the p53 tumor suppressor protein. Curr Opin Cell Biol. 2001;13:332–337. - PubMed
-
- Green DR, Chipuk JE. p53 and metabolism: Inside the TIGAR. Cell. 2006;126:30–32. - PubMed
-
- Dhanalakshmi S, Agarwal C, Singh RP, Agarwal R. Silibinin up-regulates DNA-protein kinase-dependent p53 activation to enhance UVB-induced apoptosis in mouse epithelial JB6 cells. J Biol Chem. 2005;280:20375–20383. - PubMed
-
- Reagan-Shaw S, Afaq F, Aziz MH, Ahmad N. Modulations of critical cell cycle regulatory events during chemoprevention of ultraviolet B-mediated responses by resveratrol in SKH-1 hairless mouse skin. Oncogene. 2004;23:5151–5160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

