Structure/function evaluations of single nucleotide polymorphisms in human N-acetyltransferase 2
- PMID: 18680467
- PMCID: PMC2507886
- DOI: 10.2174/138920008784892065
Structure/function evaluations of single nucleotide polymorphisms in human N-acetyltransferase 2
Abstract
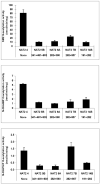
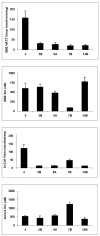
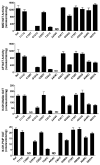
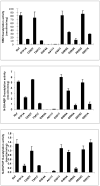
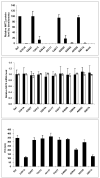
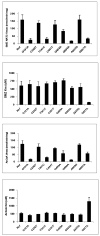
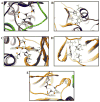
Arylamine N-acetyltransferase 2 (NAT2) modifies drug efficacy/toxicity and cancer risk due to its role in bioactivation and detoxification of arylamine and hydrazine drugs and carcinogens. Human NAT2 alleles possess a combination of single nucleotide polymorphisms (SNPs) associated with slow acetylation phenotypes. Clinical and molecular epidemiology studies investigating associations of NAT2 genotype with drug efficacy/toxicity and/or cancer risk are compromised by incomplete and sometimes conflicting information regarding genotype/phenotype relationships. Studies in our laboratory and others have characterized the functional effects of SNPs alone, and in combinations present in alleles or haplotypes. We extrapolate this data generated following recombinant expression in yeast and COS-1 cells to assist in the interpretation of NAT2 structure. Whereas previous structural studies used homology models based on templates of N-acetyltransferase enzyme crystal structures from various prokaryotic species, alignment scores between bacterial and mammalian N-acetyltransferase protein sequences are low (approximately 30%) with important differences between the bacterial and mammalian protein structures. Recently, the crystal structure of human NAT2 was released from the Protein Data Bank under accession number 2PFR. We utilized the NAT2 crystal structure to evaluate the functional effects of SNPs resulting in the protein substitutions R64Q (G191A), R64W (C190T), I114T (T341C), D122N (G364A), L137F (A411T), Q145P (A434C), E167K (G499A), R197Q (C590A), K268R (A803G), K282T (A845C), and G286E (G857A) of NAT2. This analysis advances understanding of NAT2 structure-function relationships, important for interpreting the role of NAT2 genetic polymorphisms in bioactivation and detoxification of arylamine and hydrazine drugs and carcinogens.
Figures
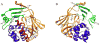

References
-
- Weber WW, Hein DW. Clin Pharmacokinet. 1979;4(6):401–422. - PubMed
-
- Weber WW, Hein DW. Pharmacol Rev. 1985;37(1):25–79. - PubMed
-
- Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y. Cancer Epidemiol Biomarkers Prev. 2000;9(1):29–42. - PubMed
-
- Hein DW. Biochim Biophys Acta. 1988;948(1):37–66. - PubMed
-
- Hein DW, Rustan TD, Doll MA, Bucher KD, Ferguson RJ, Feng Y, Furman EJ, Gray K. Toxicol Lett. 1992;64-65:123–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

