Heavily glycosylated, highly fit SIVMne variants continue to diversify and undergo selection after transmission to a new host and they elicit early antibody dependent cellular responses but delayed neutralizing antibody responses
- PMID: 18680596
- PMCID: PMC2518139
- DOI: 10.1186/1743-422X-5-90
Heavily glycosylated, highly fit SIVMne variants continue to diversify and undergo selection after transmission to a new host and they elicit early antibody dependent cellular responses but delayed neutralizing antibody responses
Abstract
Background: Lentiviruses such as human and simian immunodeficiency viruses (HIV and SIV) undergo continual evolution in the host. Previous studies showed that the late-stage variants of SIV that evolve in one host replicate to significantly higher levels when transmitted to a new host. However, it is unknown whether HIVs or SIVs that have higher replication fitness are more genetically stable upon transmission to a new host. To begin to address this, we analyzed the envelope sequence variation of viruses that evolved in animals infected with variants of SIVMne that had been cloned from an index animal at different stages of infection.
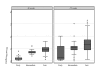
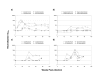
Results: We found that there was more evolution of envelope sequences from animals infected with the late-stage, highly replicating variants than in animals infected with the early-stage, lower replicating variant, despite the fact that the late virus had already diversified considerably from the early virus in the first host, prior to transmission. Many of the changes led to the addition or shift in potential-glycosylation sites-, and surprisingly, these changes emerged in some cases prior to the detection of neutralizing antibody responses, suggesting that other selection mechanisms may be important in driving virus evolution. Interestingly, these changes occurred after the development of antibody whose anti-viral function is dependent on Fc-Fcgamma receptor interactions.
Conclusion: SIV variants that had achieved high replication fitness and escape from neutralizing antibodies in one host continued to evolve upon transmission to a new host. Selection for viral variants with glycosylation and other envelope changes may have been driven by both neutralizing and Fcgamma receptor-mediated antibody activities.
Figures


References
-
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

