The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance
- PMID: 18682547
- PMCID: PMC2553615
- DOI: 10.1105/tpc.108.059444
The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance
Abstract
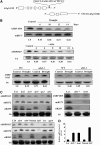
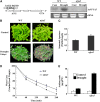
Nuclear factor Y (NF-Y) is a ubiquitous transcription factor composed of three distinct subunits (NF-YA, NF-YB, and NF-YC). We found that the Arabidopsis thaliana NFYA5 transcript is strongly induced by drought stress in an abscisic acid (ABA)-dependent manner. Promoter:beta-glucuronidase analyses showed that NFYA5 was highly expressed in vascular tissues and guard cells and that part of the induction by drought was transcriptional. NFYA5 contains a target site for miR169, which targets mRNAs for cleavage or translational repression. We found that miR169 was downregulated by drought stress through an ABA-dependent pathway. Analysis of the expression of miR169 precursors showed that miR169a and miR169c were substantially downregulated by drought stress. Coexpression of miR169 and NFYA5 suggested that miR169a was more efficient than miR169c at repressing the NFYA5 mRNA level. nfya5 knockout plants and plants overexpressing miR169a showed enhanced leaf water loss and were more sensitive to drought stress than wild-type plants. By contrast, transgenic Arabidopsis plants overexpressing NFYA5 displayed reduced leaf water loss and were more resistant to drought stress than the wild type. Microarray analysis indicated that NFYA5 is crucial for the expression of a number of drought stress-responsive genes. Thus, NFYA5 is important for drought resistance, and its induction by drought stress occurs at both the transcriptional and posttranscriptional levels.
Figures
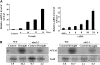
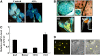


References
-
- Allen, E., Xie, Z.X., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. - PubMed
-
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. - PubMed
-
- Bohnert, H.J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms – Getting genomics going. Curr. Opin. Plant Biol. 9 180–188. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

