Transcriptional regulatory network analysis during epithelial-mesenchymal transformation of retinal pigment epithelium
- PMID: 18682805
- PMCID: PMC2493022
Transcriptional regulatory network analysis during epithelial-mesenchymal transformation of retinal pigment epithelium
Abstract
Purpose: Phenotypic transformation of retinal pigment epithelial (RPE) cells contributes to the onset and progression of ocular proliferative disorders such as proliferative vitreoretinopathy (PVR). The formation of epiretinal membranes in PVR may involve an epithelial-mesenchymal transformation (EMT) of RPE cells as part of an aberrant wound healing response. While the underlying mechanism remains unclear, this likely involves changes in RPE cell gene expression under the control of specific transcription factors (TFs). Thus, the purpose of the present study was to identify TFs that may play a role in this process.
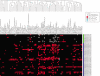
Methods: Regulatory regions of genes that are differentially regulated during phenotypic transformation of ARPE-19 cells, a human RPE cell line, were subjected to computational analysis using the promoter analysis and interaction network toolset (PAINT). The PAINT analysis was used to identify transcription response elements (TREs) statistically overrepresented in the promoter and first intron regions of two reciprocally regulated RPE gene clusters, across four species including the human genome. These TREs were then used to construct transcriptional regulatory network models of the two RPE gene clusters. The validity of these models was then tested using RT-PCR to detect differential expression of the corresponding TF mRNAs during RPE differentiation in both undifferentiated and differentiated ARPE-19 and primary chicken RPE cell cultures.
Results: The computational analysis resulted in the successful identification of specific transcription response elements (TREs) and their cognate TFs that are candidates for serving as nodes in a transcriptional regulatory network regulating EMT in RPE cells. The models predicted TFs whose differential expression during RPE EMT was successfully verified by reverse transcriptase polymerase chain reaction (RT-PCR) analysis, including Oct-1, hepatocyte nuclear factor 1 (HNF-1), similar to mothers against decapentaplegic 3 (SMAD3), transcription factor E (TFE), core binding factor, erythroid transcription factor-1 (GATA-1), interferon regulatory factor-1 (IRF), natural killer homeobox 3A (NKX3A), Sterol regulatory element binding protein-1 (SREBP-1), and lymphocyte enhancer factor-1 (LEF-1).
Conclusions: These studies successfully applied computational modeling and biochemical verification to identify biologically relevant transcription factors that are likely to regulate RPE cell phenotype and pathological changes in RPE in response to diseases or trauma. These TFs may provide potential therapeutic targets for the prevention and treatment of ocular proliferative disorders such as PVR.
Figures





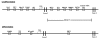







References
-
- Martinez-Morales JR, Rodrigo I, Bovolenta P. Eye development: a view from the retina pigmented epithelium. Bioessays. 2004;26:766–77. - PubMed
-
- Grunwald G. Structure and Function of the Retinal Pigment Epithelium.In: Tasman W, Jaeger EA, editors. Foundations of Clinical Ophthalmology. Philadelphia(PA): Lippincott, Williams and Wilkins; 2004. p. 1–21.
-
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–81. - PubMed
-
- Rodriguez de la Rua E, Pastor JC, Aragon J, Mayo-Iscar A, Martinez V, Garcia-Arumi J, Giraldo A, Sanabria-Ruiz Colmenares MR, Miranda I. Interaction between surgical procedure for repairing retinal detachment and clinical risk factors for proliferative vitreoretinopathy. Curr Eye Res. 2005;30:147–53. - PubMed
-
- Grierson I, Hiscott P, Hogg P, Robey H, Mazure A, Larkin G. Development, repair and regeneration of the retinal pigment epithelium. Eye. 1994;8:255–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
