Monocular visual deprivation in macaque monkeys: a profile in the gene expression of lateral geniculate nucleus by laser capture microdissection
- PMID: 18682807
- PMCID: PMC2493024
Monocular visual deprivation in macaque monkeys: a profile in the gene expression of lateral geniculate nucleus by laser capture microdissection
Abstract
Purpose: Amblyopia is the most common cause of visual impairment in children. Early detection of amblyopia and subsequent intervention are vital in preventing visual loss. Understanding the molecular pathogenesis of amblyopia would greatly facilitate development of therapeutic interventions. An animal model of amblyopia induced by monocular vision deprivation has been extensively studied in terms of anatomic and physiologic alterations that affect visual pathways. However, the molecular events underlying these changes are poorly understood. This study aimed to characterize changes of gene expression profiles in the lateral geniculate nucleus (LGN) associated with amblyopia induced by monocular visual deprivation.
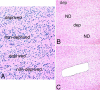
Methods: Monocular vision deprivation was generated by either opaque dark contact lens or tarsorrhaphy of newborn rhesus monkeys. LGN was harvested at two or four months following induction of vision deprivation. Laser capture microdissection was used to obtain individual LGN layers for total RNA isolation. Linear T7-based in vitro RNA amplification was used to obtain sufficient RNA to conduct DNA microarray studies. The resulting Affymetrix GeneChip Expression data were analyzed using Affymetrix GeneChip Operating Software. Real-time quantitative polymerase chain reaction and in situ hybridization were used to further analyze expression of selected genes.
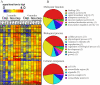
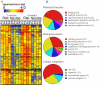
Results: Using 52,699 microarray probe sets from a Rhesus array, we identified 116 transcripts differentially expressed between deprived and nondeprived parvocellular layers: 45 genes were downregulated and 71 genes were upregulated in deprived parvocellular layers. We also observed substantial changes in deprived magnocellular laminae: 74 transcripts exhibited altered expression, 42 genes were downregulated, and 32 genes were upregulated. The genes identified in this study are involved in many diverse processes, including binding (calcium ion binding, nucleic acid binding, and nucleotide binding), catalytic activity, and signal transducer activity.
Conclusions: There were significant differences in gene expression profiles between deprived and nondeprived parvocellular layers and magnocellular laminae of LGN. These alterations in gene expression may play a critical role in the molecular pathogenesis of amblyopia. The genes identified in this study may provide potential targets for therapeutic intervention of this disease.
Figures





References
-
- von Noorden GK, Crawford ML, Levacy RA. The lateral geniculate nucleus in human anisometropic amblyopia. Invest Ophthalmol Vis Sci. 1983;24:788–90. - PubMed
-
- von Noorden GK, Crawford ML. The lateral geniculate nucleus in human strabismic amblyopia. Invest Ophthalmol Vis Sci. 1992;33:2729–32. - PubMed
-
- Daw NW. Critical periods and amblyopia. Arch Ophthalmol. 1998;116:502–5. - PubMed
-
- Headon MP, Sloper JJ, Hiorns RW, Powell TP. Effects of monocular closure at different ages on deprived and undeprived cells in the primate lateral geniculate nucleus. Brain Res. 1985;350:57–78. - PubMed
-
- Sloper JJ, Headon MP, Powell TP. Simultaneous hypertrophy of cells related to each eye in the lateral geniculate nucleus of the infant monkey following short-term reverse suture. Brain Res. 1984;317:295–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
