Pooling breast cancer datasets has a synergetic effect on classification performance and improves signature stability
- PMID: 18684329
- PMCID: PMC2527336
- DOI: 10.1186/1471-2164-9-375
Pooling breast cancer datasets has a synergetic effect on classification performance and improves signature stability
Abstract
Background: Michiels et al. (Lancet 2005; 365: 488-92) employed a resampling strategy to show that the genes identified as predictors of prognosis from resamplings of a single gene expression dataset are highly variable. The genes most frequently identified in the separate resamplings were put forward as a 'gold standard'. On a higher level, breast cancer datasets collected by different institutions can be considered as resamplings from the underlying breast cancer population. The limited overlap between published prognostic signatures confirms the trend of signature instability identified by the resampling strategy. Six breast cancer datasets, totaling 947 samples, all measured on the Affymetrix platform, are currently available. This provides a unique opportunity to employ a substantial dataset to investigate the effects of pooling datasets on classifier accuracy, signature stability and enrichment of functional categories.
Results: We show that the resampling strategy produces a suboptimal ranking of genes, which can not be considered to be a 'gold standard'. When pooling breast cancer datasets, we observed a synergetic effect on the classification performance in 73% of the cases. We also observe a significant positive correlation between the number of datasets that is pooled, the validation performance, the number of genes selected, and the enrichment of specific functional categories. In addition, we have evaluated the support for five explanations that have been postulated for the limited overlap of signatures.
Conclusion: The limited overlap of current signature genes can be attributed to small sample size. Pooling datasets results in more accurate classification and a convergence of signature genes. We therefore advocate the analysis of new data within the context of a compendium, rather than analysis in isolation.
Figures









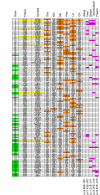
References
-
- van't Veer L, Dai H, Vijver M van de, He Y, Hart A, Mao M, Peterse H, Kooy K van der, Marton M, Witteveen A, Schreiber G, Kerhoven R, Roberts C, Linsley P, Bernards R, Friend S. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. - DOI - PubMed
-
- Wang Y, Klein J, Zhang Y, Sieuwerts A, Look M, Yang F, Talantov D, Timmermans M, Meijer-van Gelder M, Yu J, Jatkoe T, Berns E, Atkins D, Foekens J. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. - PubMed
-
- Vijver M van de, He Y, van't Veer L, Dai H, Hart A, Voskuil D, Schreiber G, Peterse J, Roberts C, Marton M, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, Velde T van der, Bartelink H, Rodenhuis S, Rutgers E, Friend S, Bernards R. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. - DOI - PubMed
-
- Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS, Bergh J, Lidereau R, Ellis P, Harris AL, Klijn JGM, Foekens JA, Cardoso F, Piccart MJ, Buyse M, Sotiriou C. Strong Time Dependence of the 76-Gene Prognostic Signature for Node-Negative Breast Cancer Patients in the TRANSBIG Multicenter Independent Validation Series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

