Transactivation of cellular genes involved in nucleotide metabolism by the regulatory IE1 protein of murine cytomegalovirus is not critical for viral replicative fitness in quiescent cells and host tissues
- PMID: 18684825
- PMCID: PMC2566287
- DOI: 10.1128/JVI.00928-08
Transactivation of cellular genes involved in nucleotide metabolism by the regulatory IE1 protein of murine cytomegalovirus is not critical for viral replicative fitness in quiescent cells and host tissues
Abstract
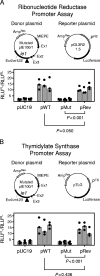
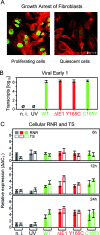
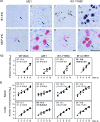
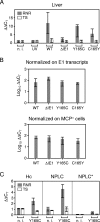
Despite its high coding capacity, murine CMV (mCMV) does not encode functional enzymes for nucleotide biosynthesis. It thus depends on cellular enzymes, such as ribonucleotide reductase (RNR) and thymidylate synthase (TS), to be supplied with deoxynucleoside triphosphates (dNTPs) for its DNA replication. Viral transactivation of these cellular genes in quiescent cells of host tissues is therefore a parameter of viral fitness relevant to pathogenicity. Previous work has shown that the IE1, but not the IE3, protein of mCMV transactivates RNR and TS gene promoters and has revealed an in vivo attenuation of the mutant virus mCMV-DeltaIE1. It was attractive to propose the hypothesis that lack of transactivation by IE1 and a resulting deficiency in the supply of dNTPs are the reasons for growth attenuation. Here, we have tested this hypothesis with the mutant virus mCMV-IE1-Y165C expressing an IE1 protein that selectively fails to transactivate RNR and TS in quiescent cells upon transfection while maintaining the capacity to disperse repressive nuclear domains (ND10). Our results confirm in vivo attenuation of mCMV-DeltaIE1, as indicated by a longer doubling time in host organs, whereas mCMV-IE1-Y165C replicated like mCMV-WT and the revertant virus mCMV-IE1-C165Y. Notably, the mutant virus transactivated RNR and TS upon infection of quiescent cells, thus indicating that IE1 is not the only viral transactivator involved. We conclude that transactivation of cellular genes of dNTP biosynthesis is ensured by redundancy and that attenuation of mCMV-DeltaIE1 results from the loss of other critical functions of IE1, with its function in the dispersal of ND10 being a promising candidate.
Figures



References
-
- Adachi, N., and M. R. Lieber. 2002. Bidirectional gene organization: a common architectural feature of the human genome. Cell 109807-809. - PubMed
-
- Applied Biosystems. 2001. User bulletin no. 2: ABI Prism 7700 Sequence Detection System (manual no. 10/2001). Applied Biosystems, Foster City, CA.
-
- Bain, M., M. Reeves, and J. Sinclair. 2006. Regulation of human cytomegalovirus gene expression by chromatin remodeling, p. 167-183. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

