Tetrahymena thermophila and Candida albicans group I intron-derived ribozymes can catalyze the trans-excision-splicing reaction
- PMID: 18684993
- PMCID: PMC2532722
- DOI: 10.1093/nar/gkn507
Tetrahymena thermophila and Candida albicans group I intron-derived ribozymes can catalyze the trans-excision-splicing reaction
Abstract
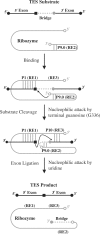
Group I intron-derived ribozymes can catalyze a variety of non-native reactions. For the trans-excision-splicing (TES) reaction, an intron-derived ribozyme from the opportunistic pathogen Pneumocystis carinii catalyzes the excision of a predefined region from within an RNA substrate with subsequent ligation of the flanking regions. To establish TES as a general ribozyme-mediated reaction, intron-derived ribozymes from Tetrahymena thermophila and Candida albicans, which are similar to but not the same as that from Pneumocystis, were investigated for their propensity to catalyze the TES reaction. We now report that the Tetrahymena and Candida ribozymes can catalyze the excision of a single nucleotide from within their ribozyme-specific substrates. Under the conditions studied, the Tetrahymena and Candida ribozymes, however, catalyze the TES reaction with lower yields and rates [Tetrahymena (k(obs)) = 0.14/min and Candida (k(obs)) = 0.34/min] than the Pneumocystis ribozyme (k(obs) = 3.2/min). The lower yields are likely partially due to the fact that the Tetrahymena and Candida catalyze additional reactions, separate from TES. The differences in rates are likely partially due to the individual ribozymes ability to effectively bind their 3' terminal guanosines as intramolecular nucleophiles. Nevertheless, our results demonstrate that group I intron-derived ribozymes are inherently able to catalyze the TES reaction.
Figures




References
-
- Cech TR. Self-splicing of group I introns. Annu. Rev. Biochem. 1990;59:543–568. - PubMed
-
- Dotson PP, II, Testa SM. Group I intron-derived ribozyme recombination reactions. Rec. Develop. Nucleic Acids Res. 2006;2:307–324.
-
- Bell MA, Johnson AJ, Testa SM. Ribozyme-catalyzed excision of targeted sequences from within RNAs. Biochemistry. 2002;41:15327–15333. - PubMed
-
- Dotson PP, II, Sinha J, Testa SM. A Pneumocystis carinii group I intron-derived ribozyme utilizes an endogenous guanosine as the first reaction step nucleophile in the trans excision-splicing reaction. Biochemistry. 2008;47:4780–4787. - PubMed
-
- Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. - PubMed

