Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro
- PMID: 18685103
- PMCID: PMC2516254
- DOI: 10.1073/pnas.0805399105
Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro
Abstract
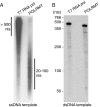
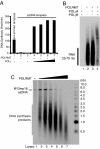
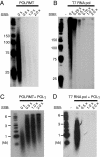
The mitochondrial transcription machinery synthesizes the RNA primers required for initiation of leading-strand DNA synthesis in mammalian mitochondria. RNA primers are also required for initiation of lagging-strand DNA synthesis, but the responsible enzyme has so far remained elusive. Here, we present a series of observations that suggests that mitochondrial RNA polymerase (POLRMT) can act as lagging-strand primase in mammalian cells. POLRMT is highly processive on double-stranded DNA, but synthesizes RNA primers with a length of 25 to 75 nt on a single-stranded template. The short RNA primers synthesized by POLRMT are used by the mitochondrial DNA polymerase gamma to initiate DNA synthesis in vitro. Addition of mitochondrial single-stranded DNA binding protein (mtSSB) reduces overall levels of primer synthesis, but stimulates primer-dependent DNA synthesis. Furthermore, when combined, POLRMT, DNA polymerase gamma, the DNA helicase TWINKLE, and mtSSB are capable of simultaneous leading- and lagging-strand DNA synthesis in vitro. Based on our observations, we suggest that POLRMT is the lagging-strand primase in mammalian mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

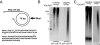
References
-
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. - PubMed
-
- Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J Biol Chem. 1981;256:5109–5115. - PubMed
-
- Kang D, Miyako K, Kai Y, Irie T, Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J Biol Chem. 1997;272:15275–15279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

