Use of quantitative pharmacology in the development of HAE1, a high-affinity anti-IgE monoclonal antibody
- PMID: 18686041
- PMCID: PMC2751395
- DOI: 10.1208/s12248-008-9045-4
Use of quantitative pharmacology in the development of HAE1, a high-affinity anti-IgE monoclonal antibody
Abstract
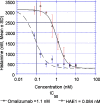
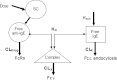
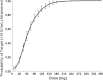
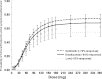
HAE1, a high-affinity anti-IgE monoclonal antibody, is discussed here as a case study in the use of quantitative pharmacology in the development of a second-generation molecule. In vitro, preclinical, and clinical data from the first-generation molecule, omalizumab, were heavily leveraged in the HAE1 program. A preliminary mechanism-based pharmacokinetic/pharmacodynamic (PK/PD) model for HAE1 was developed using an existing model for omalizumab, together with in vitro binding data for HAE1 and omalizumab. When phase I data were available, the model was refined by simultaneously modeling PK/PD data from omalizumab studies with the available HAE1 phase I data. The HAE1 clinical program was based on knowledge of the quantitative relationship between a pharmacodynamic biomarker, suppression of free IgE, and clinical response (e.g., lower exacerbation rates) obtained in pivotal studies with omalizumab. A clinical trial simulation platform was developed to predict free IgE levels and clinical responses following attainment of a target free IgE level (</=10 IU/ml). The simulation platform enabled selection of four doses for the phase II dose-ranging trial by two independent methods: dose-response non-linear fitting and linear mixed modeling. Agreement between the two methods provided confidence in the doses selected. Modeling and simulation played a large role in supporting acceleration of the HAE1 program by enabling data-driven decision-making, often based on confirmation of projections and/or learning from incoming new data.
Figures

References
-
- Ette E. I., Williams P. J. Pharmacometrics: The Science of Quantitative Pharmacology. New Jersey: Wiley; 2007.
-
- Bonate P. L. Pharmacokinetic-Pharmacodynamic Modeling and Simulation. New York: Springer Science; 2006.
-
- Lalonde R. L., Kowalski K. G., Hutmacher M. M., Ewy W., Nichols D. J., Milligan P. A., Corrigan B. W., Lockwood P. A., A Marshall S., Benincosa L. J., Tensfeldt T. G., Parivar K., Amantea M., Glue P., Koide H., Miller R. Model-based drug development. Clin. Pharmacol. Ther. 2007;82:21–32. doi: 10.1038/sj.clpt.6100235. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

