High-resolution mapping of gene expression using association in an outbred mouse stock
- PMID: 18688273
- PMCID: PMC2483929
- DOI: 10.1371/journal.pgen.1000149
High-resolution mapping of gene expression using association in an outbred mouse stock
Abstract
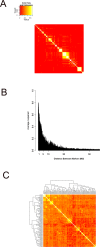
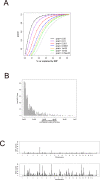
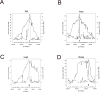
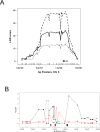
Quantitative trait locus (QTL) analysis is a powerful tool for mapping genes for complex traits in mice, but its utility is limited by poor resolution. A promising mapping approach is association analysis in outbred stocks or different inbred strains. As a proof of concept for the association approach, we applied whole-genome association analysis to hepatic gene expression traits in an outbred mouse population, the MF1 stock, and replicated expression QTL (eQTL) identified in previous studies of F2 intercross mice. We found that the mapping resolution of these eQTL was significantly greater in the outbred population. Through an example, we also showed how this precise mapping can be used to resolve previously identified loci (in intercross studies), which affect many different transcript levels (known as eQTL "hotspots"), into distinct regions. Our results also highlight the importance of correcting for population structure in whole-genome association studies in the outbred stock.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Cervino AC, Li G, Edwards S, Zhu J, Laurie C, et al. Integrating QTL and high-density SNP analyses in mice to identify Insig2 as a susceptibility gene for plasma cholesterol levels. Genomics. 2005;86:505–517. - PubMed
-
- Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol. 2000;1:221–226. - PubMed
-
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

