Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis
- PMID: 18689473
- PMCID: PMC2566193
- DOI: 10.1128/JB.00590-08
Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis
Abstract
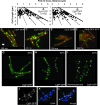
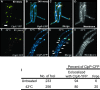
Among other functions, ATP-dependent proteases degrade misfolded proteins and remove several key regulatory proteins necessary to activate stress responses. In Bacillus subtilis, ClpX, ClpE, and ClpC form homohexameric ATPases that couple to the ClpP peptidase. To understand where these peptidases and ATPases localize in living cells, each protein was fused to a fluorescent moiety. We found that ClpX-GFP (green fluorescent protein) and ClpP-GFP localized as focal assemblies in areas that were not occupied by the nucleoid. We found that the percentage of cells with ClpP-GFP foci increased following heat shock independently of protein synthesis. We determined that ClpE-YFP (yellow fluorescent protein) and ClpC-YFP formed foci coincident with nucleoid edges, usually near cell poles. Furthermore, we found that ClpQ-YFP (HslV) localized as small foci, usually positioned near the cell membrane. We found that ClpQ-YFP foci were dependent on the presence of the cognate hexameric ATPase ClpY (HslU). Moreover, we found that LonA-GFP is coincident with the nucleoid during normal growth and that LonA-GFP also localized to the forespore during development. We also investigated LonB-GFP and found that this protein localized to the forespore membrane early in development, followed by localization throughout the forespore later in development. Our comprehensive study has shown that in B. subtilis several ATP-fueled proteases occupy distinct subcellular locations. With these data, we suggest that substrate specificity could be determined, in part, by the spatial and temporal organization of proteases in vivo.
Figures




Similar articles
-
Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis.J Bacteriol. 2008 Oct;190(20):6749-57. doi: 10.1128/JB.00589-08. Epub 2008 Aug 8. J Bacteriol. 2008. PMID: 18689476 Free PMC article.
-
Forespore-specific transcription of the lonB gene during sporulation in Bacillus subtilis.J Bacteriol. 2001 May;183(10):2995-3003. doi: 10.1128/JB.183.10.2995-3003.2001. J Bacteriol. 2001. PMID: 11325926 Free PMC article.
-
The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins.J Bacteriol. 2000 Jun;182(11):3259-65. doi: 10.1128/JB.182.11.3259-3265.2000. J Bacteriol. 2000. PMID: 10809708 Free PMC article.
-
Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation.Res Microbiol. 2004 Nov;155(9):710-9. doi: 10.1016/j.resmic.2004.06.003. Res Microbiol. 2004. PMID: 15501647 Review.
-
Chaperone-protease systems in regulation and protein quality control in Bacillus subtilis.Res Microbiol. 2009 Nov;160(9):637-44. doi: 10.1016/j.resmic.2009.08.020. Epub 2009 Sep 23. Res Microbiol. 2009. PMID: 19781636 Review.
Cited by
-
Segregation of molecules at cell division reveals native protein localization.Nat Methods. 2012 Apr 8;9(5):480-2. doi: 10.1038/nmeth.1955. Nat Methods. 2012. PMID: 22484850 Free PMC article.
-
ClpB1 overproduction in Synechocystis sp. strain PCC 6803 increases tolerance to rapid heat shock.Appl Environ Microbiol. 2013 Oct;79(20):6220-7. doi: 10.1128/AEM.01661-13. Epub 2013 Aug 2. Appl Environ Microbiol. 2013. PMID: 23913426 Free PMC article.
-
The trpE gene negatively regulates differentiation of heterocysts at the level of induction in Anabaena sp. strain PCC 7120.J Bacteriol. 2015 Jan;197(2):362-70. doi: 10.1128/JB.02145-14. Epub 2014 Nov 10. J Bacteriol. 2015. PMID: 25384479 Free PMC article.
-
Staphylococcus aureus ClpX localizes at the division septum and impacts transcription of genes involved in cell division, T7-secretion, and SaPI5-excision.Sci Rep. 2019 Nov 11;9(1):16456. doi: 10.1038/s41598-019-52823-0. Sci Rep. 2019. PMID: 31712583 Free PMC article.
-
Effect of lon Protease Overexpression on Endotoxin Production and Stress Resistance in Bacillus thuringiensis.Curr Microbiol. 2021 Sep;78(9):3483-3493. doi: 10.1007/s00284-021-02610-w. Epub 2021 Jul 17. Curr Microbiol. 2021. PMID: 34272975
References
-
- Berkmen, M. B., and A. D. Grossman. 2006. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol. Microbiol. 6257-71. - PubMed
-
- Bochtler, M., C. Hartmann, H. K. Song, G. P. Bourenkov, H. D. Bartunik, and R. Huber. 2000. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403800-805. - PubMed
-
- Bolhuis, A., A. Matzen, H. L. Hyyrylainen, V. P. Kontinen, R. Meima, J. Chapuis, G. Venema, S. Bron, R. Freudl, and J. M. van Dijl. 1999. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J. Biol. Chem. 27424585-24592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

