The amt gene cluster of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120
- PMID: 18689479
- PMCID: PMC2566009
- DOI: 10.1128/JB.00613-08
The amt gene cluster of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120
Abstract
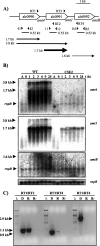
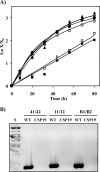
The genome of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120 bears a gene cluster including three amt genes that, based on homology of their protein products, we designate amt4, amt1, and amtB. Expression of the three genes took place upon ammonium withdrawal in combined nitrogen-free medium and was NtcA dependent. The genes were transcribed independently, but an amt4-amt1 dicistronic transcript was also produced, and expression was highest for the amt1 gene. A mutant with the whole amt region removed could grow under laboratory conditions using ammonium, nitrate, or dinitrogen as the nitrogen source.
Figures

Similar articles
-
Negative regulation of expression of the nitrate assimilation nirA operon in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120.J Bacteriol. 2010 Jun;192(11):2769-78. doi: 10.1128/JB.01668-09. Epub 2010 Mar 26. J Bacteriol. 2010. PMID: 20348260 Free PMC article.
-
Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors.Mol Microbiol. 2000 Nov;38(3):613-25. doi: 10.1046/j.1365-2958.2000.02156.x. Mol Microbiol. 2000. PMID: 11069684
-
Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120.Mol Microbiol. 1994 Nov;14(4):823-32. doi: 10.1111/j.1365-2958.1994.tb01318.x. Mol Microbiol. 1994. PMID: 7534371
-
Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development.J Bacteriol. 1994 Aug;176(15):4473-82. doi: 10.1128/jb.176.15.4473-4482.1994. J Bacteriol. 1994. PMID: 7913926 Free PMC article.
-
ATP-binding cassette transporters of the multicellular cyanobacterium Anabaena sp. PCC 7120: a wide variety for a complex lifestyle.FEMS Microbiol Lett. 2018 Feb 1;365(4). doi: 10.1093/femsle/fny012. FEMS Microbiol Lett. 2018. PMID: 29360977 Review.
Cited by
-
Complete Genome Sequence of Lactobacillus hilgardii LMG 7934, Carrying the Gene Encoding for the Novel PII-Like Protein PotN.Curr Microbiol. 2020 Nov;77(11):3538-3545. doi: 10.1007/s00284-020-02161-6. Epub 2020 Aug 14. Curr Microbiol. 2020. PMID: 32803419
-
Multiplicity and specificity of siderophore uptake in the cyanobacterium Anabaena sp. PCC 7120.Plant Mol Biol. 2016 Sep;92(1-2):57-69. doi: 10.1007/s11103-016-0495-2. Epub 2016 Jun 20. Plant Mol Biol. 2016. PMID: 27325117
-
Genomic Changes Associated with the Evolutionary Transitions of Nostoc to a Plant Symbiont.Mol Biol Evol. 2018 May 1;35(5):1160-1175. doi: 10.1093/molbev/msy029. Mol Biol Evol. 2018. PMID: 29554291 Free PMC article.
-
ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium.BMC Genomics. 2014 Jan 13;15:22. doi: 10.1186/1471-2164-15-22. BMC Genomics. 2014. PMID: 24417914 Free PMC article.
-
Coexistence of Communicating and Noncommunicating Cells in the Filamentous Cyanobacterium Anabaena.mSphere. 2021 Jan 13;6(1):e01091-20. doi: 10.1128/mSphere.01091-20. mSphere. 2021. PMID: 33441411 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2007. Current protocols in molecular biology. Greene/Wiley Interscience, New York, NY.
-
- Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 977-84. - PubMed
-
- Curatti, L., E. Flores, and G. Salerno. 2002. Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 513175-178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

