The growth-promoting and stress response activities of the Bacillus subtilis GTP binding protein Obg are separable by mutation
- PMID: 18689482
- PMCID: PMC2566217
- DOI: 10.1128/JB.00799-08
The growth-promoting and stress response activities of the Bacillus subtilis GTP binding protein Obg are separable by mutation
Abstract
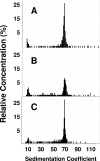
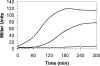
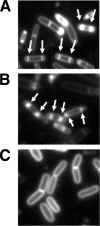
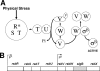
Bacillus subtilis Obg is a ribosome-associating GTP binding protein that is needed for growth, sporulation, and induction of the bacterium's general stress regulon (GSR). It is unclear whether the roles of Obg in sporulation and stress responsiveness are direct or a secondary effect of its growth-promoting functions. The present work addresses this question by an analysis of two obg alleles whose phenotypes argue for direct roles for Obg in each process. The first allele [obg(G92D)] encodes a missense change in the protein's highly conserved "obg fold" region. This mutation impairs cell growth and the ability of Obg to associate with ribosomes but fails to block sporulation or the induction of the GSR. The second obg mutation [obg(Delta22)] replaces the 22-amino-acid carboxy-terminal sequence of Obg with an alternative 26-amino-acid sequence. This Obg variant cofractionates with ribosomes and allows normal growth but blocks sporulation and impairs the induction of the GSR. Additional experiments revealed that the block on sporulation occurs early, preventing the activation of the essential sporulation transcription factor Spo0A, while inhibition of the GSR appears to involve a failure of the protein cascade that normally activates the GSR to effectively catalyze the reactions needed to activate the GSR transcription factor (sigma(B)).
Figures


Similar articles
-
Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg.J Bacteriol. 1994 Dec;176(23):7155-60. doi: 10.1128/jb.176.23.7155-7160.1994. J Bacteriol. 1994. PMID: 7961486 Free PMC article.
-
Guanine nucleotides stabilize the binding of Bacillus subtilis Obg to ribosomes.Biochem Biophys Res Commun. 2004 Sep 17;322(2):565-9. doi: 10.1016/j.bbrc.2004.07.154. Biochem Biophys Res Commun. 2004. PMID: 15325267
-
Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis.J Bacteriol. 1995 Jun;177(11):3308-11. doi: 10.1128/jb.177.11.3308-3311.1995. J Bacteriol. 1995. PMID: 7768831 Free PMC article.
-
Signal transduction in Bacillus subtilis sporulation.Curr Opin Genet Dev. 1993 Apr;3(2):203-12. doi: 10.1016/0959-437x(93)90024-j. Curr Opin Genet Dev. 1993. PMID: 8504245 Review.
-
Sporulation of Bacillus subtilis.Curr Opin Microbiol. 2004 Dec;7(6):579-86. doi: 10.1016/j.mib.2004.10.001. Curr Opin Microbiol. 2004. PMID: 15556029 Review.
Cited by
-
Simulations of stressosome activation emphasize allosteric interactions between RsbR and RsbT.BMC Syst Biol. 2013 Jan 15;7:3. doi: 10.1186/1752-0509-7-3. BMC Syst Biol. 2013. PMID: 23320651 Free PMC article.
-
STAS domain structure and function.Cell Physiol Biochem. 2011;28(3):407-22. doi: 10.1159/000335104. Epub 2011 Nov 16. Cell Physiol Biochem. 2011. PMID: 22116355 Free PMC article. Review.
-
The bacterial translation stress response.FEMS Microbiol Rev. 2014 Nov;38(6):1172-201. doi: 10.1111/1574-6976.12083. Epub 2014 Sep 26. FEMS Microbiol Rev. 2014. PMID: 25135187 Free PMC article. Review.
-
The universally conserved prokaryotic GTPases.Microbiol Mol Biol Rev. 2011 Sep;75(3):507-42, second and third pages of table of contents. doi: 10.1128/MMBR.00009-11. Microbiol Mol Biol Rev. 2011. PMID: 21885683 Free PMC article. Review.
-
The Universally Conserved ATPase YchF Regulates Translation of Leaderless mRNA in Response to Stress Conditions.Front Mol Biosci. 2021 May 7;8:643696. doi: 10.3389/fmolb.2021.643696. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026826 Free PMC article.
References
-
- Akbar, S., C. M. Kang, T. A. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24567-578. - PubMed
-
- Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260165-177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

