Regulation of the Rhodobacter sphaeroides 2.4.1 hemA gene by PrrA and FnrL
- PMID: 18689483
- PMCID: PMC2566205
- DOI: 10.1128/JB.00828-08
Regulation of the Rhodobacter sphaeroides 2.4.1 hemA gene by PrrA and FnrL
Abstract
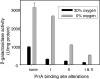
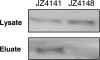
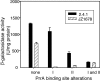
Part of the oxygen responsiveness of Rhodobacter sphaeroides 2.4.1 tetrapyrrole production involves changes in transcription of the hemA gene, which codes for one of two isoenzymes catalyzing 5-aminolevulinic acid synthesis. Regulation of hemA transcription from its two promoters is mediated by the DNA binding proteins FnrL and PrrA. The two PrrA binding sites, binding sites I and II, which are located upstream of the more-5' hemA promoter (P1), are equally important to transcription under aerobic conditions, while binding site II is more important under anaerobic conditions. By using phosphoprotein affinity chromatography and immunoblot analyses, we showed that the phosphorylated PrrA levels in the cell increase with decreasing oxygen tensions. Then, using both in vivo and in vitro methods, we demonstrated that the relative affinities of phosphorylated and unphosphorylated PrrA for the two binding sites differ and that phosphorylated PrrA has greater affinity for site II. We also showed that PrrA regulation is directed toward the P1 promoter. We propose that the PrrA component of anaerobic induction of P1 transcription is attributable to higher affinity of phosphorylated PrrA than of unphosphorylated PrrA for binding site II. Anaerobic activation of the more-3' hemA promoter (P2) is thought to involve FnrL binding to an FNR consensuslike sequence located upstream of the P2 promoter, but the contribution of FnrL to P1 induction may be indirect since the P1 transcription start is within the putative FnrL binding site. We present evidence suggesting that the indirect action of FnrL works through PrrA and discuss possible mechanisms.
Figures





Similar articles
-
Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene.J Bacteriol. 1995 Nov;177(22):6422-31. doi: 10.1128/jb.177.22.6422-6431.1995. J Bacteriol. 1995. PMID: 7592416 Free PMC article.
-
In vitro and in vivo analysis of the role of PrrA in Rhodobacter sphaeroides 2.4.1 hemA gene expression.J Bacteriol. 2006 May;188(9):3208-18. doi: 10.1128/JB.188.9.3208-3218.2006. J Bacteriol. 2006. PMID: 16621813 Free PMC article.
-
Role of the fnrL gene in photosystem gene expression and photosynthetic growth of Rhodobacter sphaeroides 2.4.1.J Bacteriol. 1998 Mar;180(6):1496-503. doi: 10.1128/JB.180.6.1496-1503.1998. J Bacteriol. 1998. PMID: 9515919 Free PMC article.
-
Control of hemA expression in Rhodobacter sphaeroides 2.4.1: regulation through alterations in the cellular redox state.J Bacteriol. 1996 Feb;178(4):985-93. doi: 10.1128/jb.178.4.985-993.1996. J Bacteriol. 1996. PMID: 8576072 Free PMC article.
-
Generalized approach to the regulation and integration of gene expression.Mol Microbiol. 2001 Mar;39(5):1116-23. doi: 10.1111/j.1365-2958.2001.02299.x. Mol Microbiol. 2001. PMID: 11251830 Review.
Cited by
-
Physiological roles for two periplasmic nitrate reductases in Rhodobacter sphaeroides 2.4.3 (ATCC 17025).J Bacteriol. 2011 Dec;193(23):6483-9. doi: 10.1128/JB.05324-11. Epub 2011 Sep 23. J Bacteriol. 2011. PMID: 21949073 Free PMC article.
-
A New Strategy for Production of 5-Aminolevulinic Acid in Recombinant Corynebacterium glutamicum with High Yield.Appl Environ Microbiol. 2016 Apr 18;82(9):2709-2717. doi: 10.1128/AEM.00224-16. Print 2016 May. Appl Environ Microbiol. 2016. PMID: 26921424 Free PMC article.
-
Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control.Antioxid Redox Signal. 2012 Apr 15;16(8):819-52. doi: 10.1089/ars.2011.4051. Epub 2012 Jan 25. Antioxid Redox Signal. 2012. PMID: 22098259 Free PMC article. Review.
-
The Global Redox Responding RegB/RegA Signal Transduction System Regulates the Genes Involved in Ferrous Iron and Inorganic Sulfur Compound Oxidation of the Acidophilic Acidithiobacillus ferrooxidans.Front Microbiol. 2017 Jul 12;8:1277. doi: 10.3389/fmicb.2017.01277. eCollection 2017. Front Microbiol. 2017. PMID: 28747899 Free PMC article.
-
CbbR, the Master Regulator for Microbial Carbon Dioxide Fixation.J Bacteriol. 2015 Nov;197(22):3488-98. doi: 10.1128/JB.00442-15. Epub 2015 Aug 31. J Bacteriol. 2015. PMID: 26324454 Free PMC article. Review.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
-
- Barber, R. D., and T. J. Donohue. 1998. Pathways for transcriptional activation of a glutathione-dependent formaldehyde dehydrogenase gene. J. Mol. Biol. 280775-784. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

