Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR
- PMID: 18689485
- PMCID: PMC2580704
- DOI: 10.1128/JB.00855-08
Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR
Abstract
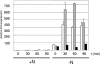
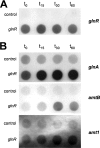
The effect of nitrogen regulation on the level of transcriptional control has been investigated in a variety of bacteria, such as Bacillus subtilis, Corynebacterium glutamicum, Escherichia coli, and Streptomyces coelicolor; however, until now there have been no data for mycobacteria. In this study, we found that the OmpR-type regulator protein GlnR controls nitrogen-dependent transcription regulation in Mycobacterium smegmatis. Based on RNA hybridization experiments with a wild-type strain and a corresponding mutant strain, real-time reverse transcription-PCR analyses, and DNA binding studies using cell extract and purified protein, the glnA (msmeg_4290) gene, which codes for glutamine synthetase, and the amtB (msmeg_2425) and amt1 (msmeg_6259) genes, which encode ammonium permeases, are controlled by GlnR. Furthermore, since glnK (msmeg_2426), encoding a PII-type signal transduction protein, and glnD (msmeg_2427), coding for a putative uridylyltransferase, are in an operon together with amtB, these genes are part of the GlnR regulon as well. The GlnR protein binds specifically to the corresponding promoter sequences and functions as an activator of transcription when cells are subjected to nitrogen starvation.
Figures






References
-
- Beckers, G., J. Strösser, U. Hildebrandt, J. Kalinowski, M. Farwick, R. Krämer, and A. Burkovski. 2005. Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: mechanism and characterization of the AmtR regulon. Mol. Microbiol. 58580-595. - PubMed
-
- Belisle, J. T., and M. G. Sonnenberg. 1998. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 10131-44. - PubMed
-
- Burkovski, A. 2003. Ammonium assimilation and nitrogen control in Corynebacterium glutamicum and its relatives: an example for new regulatory mechanisms in actinomycetes. FEMS Microbiol. Rev. 27617-628. - PubMed
-
- Burkovski, A. 2005. Nitrogen metabolism and its regulation, p. 333-349. In M. Bott and L. Eggeling (ed.), Handbook of Corynebacterium glutamicum. CRC Press LLC, Boca Raton, FL.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

