Identification of residues important for cleavage of the extracellular signaling peptide CSF of Bacillus subtilis from its precursor protein
- PMID: 18689487
- PMCID: PMC2566178
- DOI: 10.1128/JB.00910-08
Identification of residues important for cleavage of the extracellular signaling peptide CSF of Bacillus subtilis from its precursor protein
Abstract
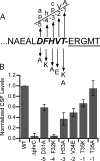
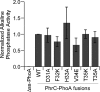
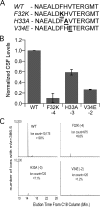
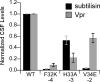
Extracellular Phr pentapeptides produced by gram-positive, spore-forming bacteria regulate processes during the transition from exponential- to stationary-phase growth. Phr pentapeptides are produced by cleavage of their precursor proteins. We determined the residues that direct this cleavage for the Bacillus subtilis Phr peptide, CSF, which is derived from the C terminus of PhrC. Strains expressing PhrC with substitutions in residues -1 to -5 relative to the cleavage site had a defect in CSF production. The mutant PhrC proteins retained a functional signal sequence for secretion, as assessed by secretion of PhrC-PhoA fusions. To determine whether the substitutions directly affected cleavage of PhrC to CSF, we tested cleavage of synthetic pro-CSF peptides that corresponded to the C terminus of PhrC and had an amino acid substitution at the -2, -3, or -4 position. The mutant pro-CSF peptides were cleaved less efficiently to CSF than the wild-type pro-CSF peptide whether they were incubated with whole cells, cell wall material, or the processing protease subtilisin or Vpr. To further define the range of amino acids that support CSF production, the amino acid at the -4 position of PhrC was replaced by the 19 canonical amino acids. Only four substitutions resulted in a >2-fold defect in CSF production, indicating that this position is relatively immune to mutational perturbations. These data revealed residues that direct cleavage of CSF and laid the groundwork for testing whether other Phr peptides are processed in a similar manner.
Figures
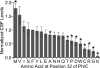
Similar articles
-
Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis.Mol Microbiol. 2007 Sep;65(5):1321-33. doi: 10.1111/j.1365-2958.2007.05869.x. Epub 2007 Jul 31. Mol Microbiol. 2007. PMID: 17666034
-
Use of alkaline phosphatase as a reporter polypeptide to study the role of the subtilin leader segment and the SpaT transporter in the posttranslational modifications and secretion of subtilin in Bacillus subtilis 168.Appl Environ Microbiol. 1997 Oct;63(10):3965-71. doi: 10.1128/aem.63.10.3965-3971.1997. Appl Environ Microbiol. 1997. PMID: 9327561 Free PMC article.
-
An autoregulatory circuit affecting peptide signaling in Bacillus subtilis.J Bacteriol. 1999 Sep;181(17):5193-200. doi: 10.1128/JB.181.17.5193-5200.1999. J Bacteriol. 1999. PMID: 10464187 Free PMC article.
-
Molecular analysis of Phr peptide processing in Bacillus subtilis.J Bacteriol. 2003 Aug;185(16):4861-71. doi: 10.1128/JB.185.16.4861-4871.2003. J Bacteriol. 2003. PMID: 12897006 Free PMC article.
-
The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis.Front Biosci. 2003 Jan 1;8:d32-45. doi: 10.2741/913. Front Biosci. 2003. PMID: 12456319 Review.
Cited by
-
Peptide-based quorum sensing systems in Paenibacillus polymyxa.Life Sci Alliance. 2020 Aug 6;3(10):e202000847. doi: 10.26508/lsa.202000847. Print 2020 Oct. Life Sci Alliance. 2020. PMID: 32764104 Free PMC article.
-
Peptidome: Chaos or Inevitability.Int J Mol Sci. 2021 Dec 4;22(23):13128. doi: 10.3390/ijms222313128. Int J Mol Sci. 2021. PMID: 34884929 Free PMC article. Review.
-
Bacterial transformation: distribution, shared mechanisms and divergent control.Nat Rev Microbiol. 2014 Mar;12(3):181-96. doi: 10.1038/nrmicro3199. Epub 2014 Feb 10. Nat Rev Microbiol. 2014. PMID: 24509783 Review.
-
Genetic and Structural Analyses of RRNPP Intercellular Peptide Signaling of Gram-Positive Bacteria.Annu Rev Genet. 2017 Nov 27;51:311-333. doi: 10.1146/annurev-genet-120116-023507. Epub 2017 Sep 6. Annu Rev Genet. 2017. PMID: 28876981 Free PMC article. Review.
-
Osmoprotection of Bacillus subtilis through import and proteolysis of proline-containing peptides.Appl Environ Microbiol. 2013 Jan;79(2):576-87. doi: 10.1128/AEM.01934-12. Epub 2012 Nov 9. Appl Environ Microbiol. 2013. PMID: 23144141 Free PMC article.
References
-
- Bassler, B., P. Gibbons, and S. Roseman. 1989. Chemotaxis to chitin oligosaccharides by Vibrio furnissii, a chitinivorous marine bacterium. Biochem. Biophys. Res. Commun. 1611172-1176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

