Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation
- PMID: 18689544
- PMCID: PMC2572803
- DOI: 10.1182/blood-2008-04-154286
Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation
Abstract
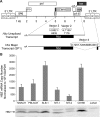
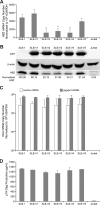
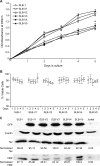
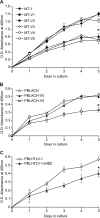
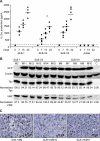
Human T-cell leukemia virus type 1 (HTLV-1) basic leucine zipper factor (HBZ) is dispensable for HTLV-1-mediated cellular transformation in cell culture, but is required for efficient viral infectivity and persistence in rabbits. In most adult T-cell leukemia (ATL) cells, Tax oncoprotein expression is typically low or undetectable, whereas Hbz gene expression is maintained, suggesting that Hbz expression may support infected cell survival and, ultimately, leukemogenesis. Emerging data indicate that HBZ protein can interact with cAMP response element binding protein (CREB) and Jun family members, altering transcription factor binding and transactivation of both viral and cellular promoters. Herein, lentiviral vectors that express Hbz-specific short hairpin (sh)-RNA effectively decreased both Hbz mRNA and HBZ protein expression in transduced HTLV-1-transformed SLB-1 T cells. Hbz knockdown correlated with a significant decrease in T-cell proliferation in culture. Both SLB-1 and SLB-1-Hbz knockdown cells engrafted into inoculated NOD/SCID(gammachain-/-) mice to form solid tumors that also infiltrated multiple tissues. However, tumor formation and organ infiltration were significantly decreased in animals challenged with SLB-1-Hbz knockdown cells. Our data indicate that Hbz expression enhances the proliferative capacity of HTLV-1-infected T cells, playing a critical role in cell survival and ultimately HTLV-1 tumorigenesis in the infected host.
Figures

References
-
- Lairmore M, Franchini G. Human T-cell leukemia virus types 1 and 2. In: Fields B, Knipe D, Howley P, et al., editors. Fields Virology. Vol. 5. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. pp. 2071–2106. Chapter 56.
-
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. - PubMed
-
- Collins ND, Newbound GC, Albrecht B, Beard J, Ratner L, Lairmore MD. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood. 1998;91:4701–4707. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

