Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal
- PMID: 18689797
- PMCID: PMC2562071
- DOI: 10.1074/jbc.M804550200
Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal
Abstract
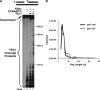
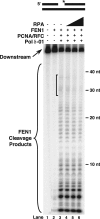
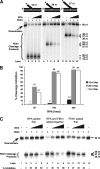
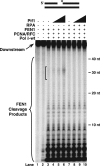
Eukaryotic Okazaki fragment maturation requires complete removal of the initiating RNA primer before ligation occurs. Polymerase delta (Pol delta) extends the upstream Okazaki fragment and displaces the 5'-end of the downstream primer into a single nucleotide flap, which is removed by FEN1 nuclease cleavage. This process is repeated until all RNA is removed. However, a small fraction of flaps escapes cleavage and grows long enough to be coated with RPA and requires the consecutive action of the Dna2 and FEN1 nucleases for processing. Here we tested whether RPA inhibits FEN1 cleavage of long flaps as proposed. Surprisingly, we determined that RPA binding to long flaps made dynamically by polymerase delta only slightly inhibited FEN1 cleavage, apparently obviating the need for Dna2. Therefore, we asked whether other relevant proteins promote long flap cleavage via the Dna2 pathway. The Pif1 helicase, implicated in Okazaki maturation from genetic studies, improved flap displacement and increased RPA inhibition of long flap cleavage by FEN1. These results suggest that Pif1 accelerates long flap growth, allowing RPA to bind before FEN1 can act, thereby inhibiting FEN1 cleavage. Therefore, Pif1 directs long flaps toward the two-nuclease pathway, requiring Dna2 cleavage for primer removal.
Figures


Similar articles
-
An alternative pathway for Okazaki fragment processing: resolution of fold-back flaps by Pif1 helicase.J Biol Chem. 2010 Dec 31;285(53):41712-23. doi: 10.1074/jbc.M110.146894. Epub 2010 Oct 19. J Biol Chem. 2010. PMID: 20959454 Free PMC article.
-
Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing.J Biol Chem. 2010 Sep 10;285(37):28496-505. doi: 10.1074/jbc.M110.131870. Epub 2010 Jul 13. J Biol Chem. 2010. PMID: 20628185 Free PMC article.
-
Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae.J Biol Chem. 2009 Mar 27;284(13):8283-91. doi: 10.1074/jbc.M809189200. Epub 2009 Jan 29. J Biol Chem. 2009. PMID: 19179330 Free PMC article.
-
Polymerase dynamics at the eukaryotic DNA replication fork.J Biol Chem. 2009 Feb 13;284(7):4041-5. doi: 10.1074/jbc.R800062200. Epub 2008 Oct 3. J Biol Chem. 2009. PMID: 18835809 Free PMC article. Review.
-
Reconstitution of eukaryotic lagging strand DNA replication.Methods. 2010 Jul;51(3):347-57. doi: 10.1016/j.ymeth.2010.02.017. Epub 2010 Feb 21. Methods. 2010. PMID: 20178844 Free PMC article. Review.
Cited by
-
Mechanistic investigation of human maturation of Okazaki fragments reveals slow kinetics.Nat Commun. 2022 Nov 15;13(1):6973. doi: 10.1038/s41467-022-34751-2. Nat Commun. 2022. PMID: 36379932 Free PMC article.
-
Pif1, RPA, and FEN1 modulate the ability of DNA polymerase δ to overcome protein barriers during DNA synthesis.J Biol Chem. 2020 Nov 20;295(47):15883-15891. doi: 10.1074/jbc.RA120.015699. Epub 2020 Sep 10. J Biol Chem. 2020. PMID: 32913126 Free PMC article.
-
TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication.J Biol Chem. 2010 Mar 5;285(10):7056-66. doi: 10.1074/jbc.M109.084038. Epub 2009 Dec 30. J Biol Chem. 2010. PMID: 20042610 Free PMC article.
-
Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint.Cell Cycle. 2011 May 15;10(10):1690-8. doi: 10.4161/cc.10.10.15643. Epub 2011 May 15. Cell Cycle. 2011. PMID: 21508669 Free PMC article.
-
Pif1-Family Helicases Support Fork Convergence during DNA Replication Termination in Eukaryotes.Mol Cell. 2019 Apr 18;74(2):231-244.e9. doi: 10.1016/j.molcel.2019.01.040. Epub 2019 Mar 5. Mol Cell. 2019. PMID: 30850330 Free PMC article.
References
-
- Kornberg, A., and Baker, T. A. (1992) in DNA Replication, 2nd Ed., pp. 113–195, W. H. Freeman, New York
-
- Bambara, R. A., Murante, R. S., and Henricksen, L. A. (1997) J. Biol. Chem. 272 4647–4650 - PubMed
-
- Liu, Y., Kao, H. I., and Bambara, R. A. (2004) Annu. Rev. Biochem. 73 589–615 - PubMed
-
- Rossi, M. L., Purohit, V., Brandt, P. D., and Bambara, R. A. (2006) Chem. Rev. 106 453–473 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

