Subdivision in an ancestral species creates asymmetry in gene trees
- PMID: 18689871
- PMCID: PMC2734134
- DOI: 10.1093/molbev/msn172
Subdivision in an ancestral species creates asymmetry in gene trees
Abstract
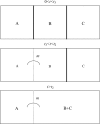
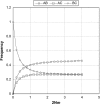
We consider gene trees in three species for which the species tree is known. We show that population subdivision in ancestral species can lead to asymmetry in the frequencies of the two gene trees not concordant with the species tree and, if subdivision is extreme, cause the one of the nonconcordant gene trees to be more probable than the concordant gene tree. Although published data for the human-chimp-gorilla clade and for three species of Drosophila show asymmetry consistent with our model, sequencing error could also account for observed patterns. We show that substantial levels of persistent ancestral subdivision are needed to account for the observed levels of asymmetry found in these two studies.
Figures


References
-
- Aquadro CF, DuMont VB, Reed FA. Genome-wide variation in the human and fruitfly: a comparison. Curr Opin Genet Dev. 2001;11:627–634. - PubMed
-
- Hudson RR. Testing the constant-rate neutral allele model with protein sequence data. Evolution. 1983;37:203–217. - PubMed
-
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

