Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish
- PMID: 18689886
- PMCID: PMC2516089
- DOI: 10.1534/genetics.108.088559
Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish
Abstract
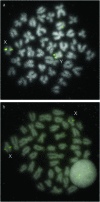
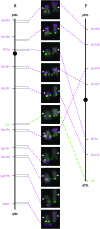
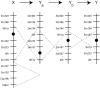
To identify the processes shaping vertebrate sex chromosomes during the early stages of their evolution, it is necessary to study systems in which genetic sex determination was recently acquired. Previous cytogenetic studies suggested that threespine stickleback fish (Gasterosteus aculeatus) do not have a heteromorphic sex chromosome pair, although recent genetic studies found evidence of an XY genetic sex-determination system. Using fluorescence in situ hybridization (FISH), we report that the threespine stickleback Y chromosome is heteromorphic and has suffered both inversions and deletion. Using the FISH data, we reconstruct the rearrangements that have led to the current physical state of the threespine stickleback Y chromosome. These data demonstrate that the threespine Y is more degenerate than previously thought, suggesting that the process of sex chromosome evolution can occur rapidly following acquisition of a sex-determining region.
Figures




Similar articles
-
Karyotype differentiation between two stickleback species (Gasterosteidae).Cytogenet Genome Res. 2011;135(2):150-9. doi: 10.1159/000331232. Epub 2011 Sep 13. Cytogenet Genome Res. 2011. PMID: 21921583 Free PMC article.
-
Assembly of the threespine stickleback Y chromosome reveals convergent signatures of sex chromosome evolution.Genome Biol. 2020 Jul 19;21(1):177. doi: 10.1186/s13059-020-02097-x. Genome Biol. 2020. PMID: 32684159 Free PMC article.
-
The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome.Curr Biol. 2004 Aug 24;14(16):1416-24. doi: 10.1016/j.cub.2004.08.030. Curr Biol. 2004. PMID: 15324658
-
Threespine Stickleback: A Model System For Evolutionary Genomics.Annu Rev Genomics Hum Genet. 2021 Aug 31;22:357-383. doi: 10.1146/annurev-genom-111720-081402. Epub 2021 Apr 28. Annu Rev Genomics Hum Genet. 2021. PMID: 33909459 Free PMC article. Review.
-
Studies of threespine stickleback developmental evolution: progress and promise.Genetica. 2007 Jan;129(1):105-26. doi: 10.1007/s10709-006-0036-z. Epub 2006 Aug 8. Genetica. 2007. PMID: 16897450 Review.
Cited by
-
Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes.Mol Biol Evol. 2013 May;30(5):1131-44. doi: 10.1093/molbev/mst035. Epub 2013 Feb 23. Mol Biol Evol. 2013. PMID: 23436913 Free PMC article.
-
Chromosomal Fusions Facilitate Adaptation to Divergent Environments in Threespine Stickleback.Mol Biol Evol. 2022 Feb 3;39(2):msab358. doi: 10.1093/molbev/msab358. Mol Biol Evol. 2022. PMID: 34908155 Free PMC article.
-
Karyotype differentiation between two stickleback species (Gasterosteidae).Cytogenet Genome Res. 2011;135(2):150-9. doi: 10.1159/000331232. Epub 2011 Sep 13. Cytogenet Genome Res. 2011. PMID: 21921583 Free PMC article.
-
The DNA Methylation Landscape of Stickleback Reveals Patterns of Sex Chromosome Evolution and Effects of Environmental Salinity.Genome Biol Evol. 2018 Mar 1;10(3):775-785. doi: 10.1093/gbe/evy034. Genome Biol Evol. 2018. PMID: 29420714 Free PMC article.
-
Genetic Architecture of Conspicuous Red Ornaments in Female Threespine Stickleback.G3 (Bethesda). 2015 Dec 29;6(3):579-88. doi: 10.1534/g3.115.024505. G3 (Bethesda). 2015. PMID: 26715094 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

