Bayesian quantitative trait loci mapping for multiple traits
- PMID: 18689903
- PMCID: PMC2516097
- DOI: 10.1534/genetics.108.088427
Bayesian quantitative trait loci mapping for multiple traits
Abstract
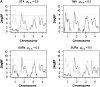
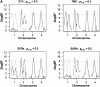
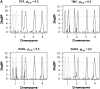
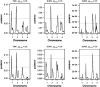
Most quantitative trait loci (QTL) mapping experiments typically collect phenotypic data on multiple correlated complex traits. However, there is a lack of a comprehensive genomewide mapping strategy for correlated traits in the literature. We develop Bayesian multiple-QTL mapping methods for correlated continuous traits using two multivariate models: one that assumes the same genetic model for all traits, the traditional multivariate model, and the other known as the seemingly unrelated regression (SUR) model that allows different genetic models for different traits. We develop computationally efficient Markov chain Monte Carlo (MCMC) algorithms for performing joint analysis. We conduct extensive simulation studies to assess the performance of the proposed methods and to compare with the conventional single-trait model. Our methods have been implemented in the freely available package R/qtlbim (http://www.qtlbim.org), which greatly facilitates the general usage of the Bayesian methodology for unraveling the genetic architecture of complex traits.
Figures



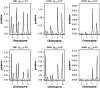
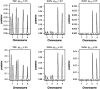
References
-
- Broman, K. W., H. Wu, Ś. Sen and G. A. Churchill, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 889–890. - PubMed
-
- Burnham, K. P., and D. R. Anderson, 2002. Model Selection and Multi-Model Inference. Springer-Verlag, New York.
-
- Gelman, A., J. Carlin, H. Stern and D. Rubin, 2004. Bayesian Data Analysis. Chapman & Hall/CRC, London.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

