Widespread impact of nonsense-mediated mRNA decay on the yeast intronome
- PMID: 18691968
- PMCID: PMC2600495
- DOI: 10.1016/j.molcel.2008.07.005
Widespread impact of nonsense-mediated mRNA decay on the yeast intronome
Abstract
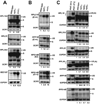
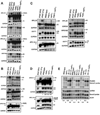
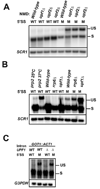
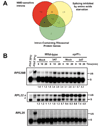
Nonsense-mediated mRNA decay (NMD) eliminates transcripts carrying premature translation termination codons, but the role of NMD on yeast unspliced pre-mRNA degradation is controversial. Using tiling arrays, we show that many unspliced yeast pre-mRNAs accumulate in strains mutated for the NMD component Upf1p and the exonuclease Xrn1p. Intron identity and suboptimal splicing signals resulting in weak splicing were found to be important determinants in NMD targeting. In the absence of functional NMD, unspliced precursors accumulate in the cytoplasm, possibly in P-bodies. NMD can also complement RNase III-mediated nuclear degradation of unspliced RPS22B pre-mRNAs, degrades most unspliced precursors generated by a 5' splice site mutation in RPS10B, and limits RPS29B unspliced precursors accumulation during amino acid starvation. These results show that NMD has a wider impact than previously thought on the degradation of yeast-unspliced transcripts and plays an important role in discarding precursors of regulated or suboptimally spliced transcripts.
Figures



References
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. - PubMed
-
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. - PubMed
-
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. - PubMed
-
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. - PubMed
-
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

