General and versatile autoinhibition of PLC isozymes
- PMID: 18691970
- PMCID: PMC2702322
- DOI: 10.1016/j.molcel.2008.06.018
General and versatile autoinhibition of PLC isozymes
Abstract
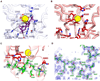
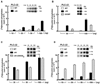
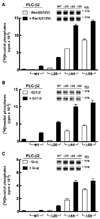
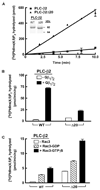
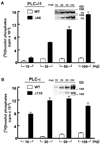
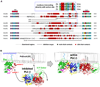
Phospholipase C (PLC) isozymes are directly activated by heterotrimeric G proteins and Ras-like GTPases to hydrolyze phosphatidylinositol 4,5-bisphosphate into the second messengers diacylglycerol and inositol 1,4,5-trisphosphate. Although PLCs play central roles in myriad signaling cascades, the molecular details of their activation remain poorly understood. As described here, the crystal structure of PLC-beta2 illustrates occlusion of the active site by a loop separating the two halves of the catalytic TIM barrel. Removal of this insertion constitutively activates PLC-beta2 without ablating its capacity to be further stimulated by classical G protein modulators. Similar regulation occurs in other PLC members, and a general mechanism of interfacial activation at membranes is presented that provides a unifying framework for PLC activation by diverse stimuli.
Figures

References
-
- Barr AJ, Ali H, Haribabu B, Snyderman R, Smrcka AV. Identification of a region at the N-terminus of phospholipase C-β3 that interacts with G protein βγ subunits. Biochemistry. 2000;39:1800–1806. - PubMed
-
- Blank JL, Shaw K, Ross AH, Exton JH. Purification of a 110-kDa phosphoinositide phospholipase C that is activated by G-protein βγsubunits. J. Biol. Chem. 1993;268:25184–25191. - PubMed
-
- Boyer JL, Waldo GL, Harden TK. βγ-subunit activation of G-protein- regulated phospholipase C. J. Biol. Chem. 1992;267:25451–25456. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. - PubMed
-
- (CCP4) Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta. Cryst. D. 1994;50:760–763. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

