GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors
- PMID: 18692040
- PMCID: PMC3031907
- DOI: 10.1016/j.ydbio.2008.07.022
GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors
Abstract
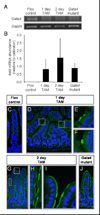
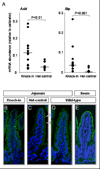
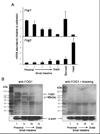
GATA4, a transcription factor expressed in the proximal small intestine but not in the distal ileum, maintains proximal-distal distinctions by multiple processes involving gene repression, gene activation, and cell fate determination. Friend of GATA (FOG) is an evolutionarily conserved family of cofactors whose members physically associate with GATA factors and mediate GATA-regulated repression in multiple tissues. Using a novel, inducible, intestine-specific Gata4 knock-in model in mice, in which wild-type GATA4 is specifically inactivated in the small intestine, but a GATA4 mutant that does not bind FOG cofactors (GATA4ki) continues to be expressed, we found that ileal-specific genes were significantly induced in the proximal small intestine (P<0.01); in contrast, genes restricted to proximal small intestine and cell lineage markers were unaffected, indicating that GATA4-FOG interactions contribute specifically to the repression function of GATA4 within this organ. Fog1 mRNA displayed a proximal-distal pattern that parallels that of Gata4, and FOG1 protein was co-expressed with GATA4 in intestinal epithelial cells, implicating FOG1 as the likely mediator of GATA4 function in the small intestine. Our data are the first to indicate FOG function and expression in the mammalian small intestine.
Figures





References
-
- Bosse T, et al. Gata4 and Hnf1alpha are partially required for the expression of specific intestinal genes during development. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1302–G1314. - PubMed
-
- Bosse T, et al. Hepatocyte nuclear factor-1alpha is required for expression but dispensable for histone acetylation of the lactase-phlorizin hydrolase gene in vivo. Am J Physiol Gastrointest Liver Physiol. 2006b;290:G1016–G1024. - PubMed
-
- Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol. 2005;16:117–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

