Characterization of structural variations in the peptidoglycan of vancomycin-susceptible Enterococcus faecium: understanding glycopeptide-antibiotic binding sites using mass spectrometry
- PMID: 18692403
- PMCID: PMC2613859
- DOI: 10.1016/j.jasms.2008.06.020
Characterization of structural variations in the peptidoglycan of vancomycin-susceptible Enterococcus faecium: understanding glycopeptide-antibiotic binding sites using mass spectrometry
Abstract
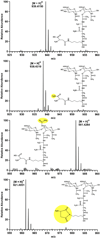
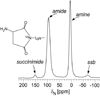
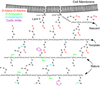
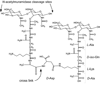
Enterococcus faecium, an opportunistic pathogen that causes a significant number of hospital-acquired infections each year, presents a serious clinical challenge because an increasing number of infections are resistant to the so-called antibiotic of last resort, vancomycin. Vancomycin and other new glycopeptide derivatives target the bacterial cell wall, thereby perturbing its biosynthesis. To help determine the modes of action of glycopeptide antibiotics, we have developed a bottom-up mass spectrometry approach complemented by solid-state nuclear magnetic resonance (NMR) to elucidate important structural characteristics of vancomycin-susceptible E. faecium peptidoglycan. Using accurate-mass measurements and integrating ion-current chromatographic peaks of digested peptidoglycan, we identified individual muropeptide species and approximated the relative amount of each. Even though the organism investigated is susceptible to vancomycin, only 3% of the digested peptidoglycan has the well-known D-Ala-D-Ala vancomycin-binding site. The data are consistent with a previously proposed template model of cell-wall biosynthesis where D-Ala-D-Ala stems that are not cross-linked are cleaved in mature peptidoglycan. Additionally, our mass-spectrometry approach allowed differentiation and quantification of muropeptide species seen as unresolved chromatographic peaks. Our method provides an estimate of the extent of muropeptides containing O-acetylation, amidation, hydroxylation, and the number of species forming cyclic imides. The varieties of muropeptides on which the modifications are detected suggest that significant processing occurs in mature peptidoglycan where several enzymes are active in editing cell-wall structure.
Figures



References
-
- Uttley AH, Collins CH, Naidoo J, George RC. Vancomycin-resistant enterococci. Lancet. 1988;1:57–58. - PubMed
-
- Kak VAC, Joseph W. Acquired antibiotic resistances in enterococci. Washington, DC: ASM Press; 2002.
-
- Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 2008;32:386–408. - PubMed
-
- Courvalin P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42. 2006;Suppl 1:S25–S34. - PubMed
-
- Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

