A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing
- PMID: 18692465
- PMCID: PMC2602844
- DOI: 10.1016/j.cell.2008.06.021
A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing
Abstract
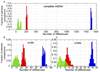
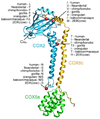
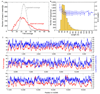
A complete mitochondrial (mt) genome sequence was reconstructed from a 38,000 year-old Neandertal individual with 8341 mtDNA sequences identified among 4.8 Gb of DNA generated from approximately 0.3 g of bone. Analysis of the assembled sequence unequivocally establishes that the Neandertal mtDNA falls outside the variation of extant human mtDNAs, and allows an estimate of the divergence date between the two mtDNA lineages of 660,000 +/- 140,000 years. Of the 13 proteins encoded in the mtDNA, subunit 2 of cytochrome c oxidase of the mitochondrial electron transport chain has experienced the largest number of amino acid substitutions in human ancestors since the separation from Neandertals. There is evidence that purifying selection in the Neandertal mtDNA was reduced compared with other primate lineages, suggesting that the effective population size of Neandertals was small.
Figures


Comment in
-
Genome sequences from extinct relatives.Cell. 2008 Aug 8;134(3):388-9. doi: 10.1016/j.cell.2008.07.026. Cell. 2008. PMID: 18692462
-
Evolutionary genomics: our Neanderthal cousins.Nat Rev Genet. 2008 Sep;9(9):652. doi: 10.1038/nrg2448. Nat Rev Genet. 2008. PMID: 21491640 No abstract available.
References
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. - PubMed
-
- Belevich I, Verkhovsky MI, Wikstrom M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature. 2006;440:829–832. - PubMed
-
- Brunet M, Guy F, Pilbeam D, Mackaye HT, Likius A, Ahounta D, Beauvilain A, Blondel C, Bocherens H, Boisserie JR, et al. A new hominid from the Upper Miocene of Chad, Central Africa. Nature. 2002;418:145–151. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

