A network of nuclear envelope membrane proteins linking centromeres to microtubules
- PMID: 18692466
- PMCID: PMC2617791
- DOI: 10.1016/j.cell.2008.06.022
A network of nuclear envelope membrane proteins linking centromeres to microtubules
Abstract
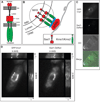
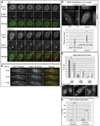
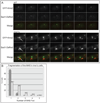
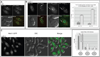
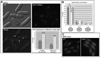
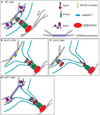
In the fission yeast S. pombe, nuclei are actively positioned at the cell center by microtubules. Here, we show that cytoplasmic microtubules are mechanically coupled to the nuclear heterochromatin through proteins embedded in the nuclear envelope. This includes an integral outer nuclear membrane protein of the KASH family (Kms2) and two integral inner nuclear membrane proteins, the SUN-domain protein Sad1 and the previously uncharacterized protein Ima1. Ima1 specifically binds to heterochromatic regions and promotes the tethering of centromeric DNA to the SUN-KASH complex. In the absence of Ima1, or in cells harboring mutations in the centromeric Ndc80 complex, inefficient coupling of centromeric heterochromatin to Sad1 leads to striking defects in the ability of the nucleus to tolerate microtubule-dependent forces, leading to changes in nuclear shape, loss of spindle pole body components from the nuclear envelope, and partial dissociation of SUN-KASH complexes. This work highlights a framework for communication between cytoplasmic microtubules and chromatin.
Figures

References
-
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. - PubMed
-
- Appelgren H, Kniola B, Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. - PubMed
-
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

