Transcriptional interference antagonizes proviral gene expression to promote HIV latency
- PMID: 18692772
- PMCID: PMC4217705
- DOI: 10.1016/j.chom.2008.05.016
Transcriptional interference antagonizes proviral gene expression to promote HIV latency
Abstract
Eradication of the latent HIV reservoir remains a major barrier to curing AIDS. However, the mechanisms that direct viral persistence in the host are not well understood. Studying a model system of postintegration latency, we found that viral integration into the actively transcribed host genes led to transcriptional interference (TI) caused by the elongating RNA polymerase II (RNAPII) transcribing through the viral promoter. The resulting physical exclusion of preinitiation complex formation on the 5' long terminal repeat (LTR) promoted the silencing of HIV transcription. This block could be counteracted by inhibiting the upstream transcription or cooperatively activating viral transcription initiation and elongation. Importantly, PCR-based analysis, which detects host transcription through the 5'LTR independently of the viral integration site, revealed substantial levels of this transcription in HIV-infected primary CD4(+) T cells. Collectively, our findings suggest that TI contributes significantly to HIV latency and should be considered when attempting to purge the latent reservoir.
Figures

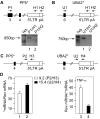
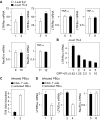
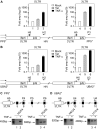


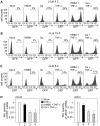
Comment in
-
An ungracious host for an unwelcome guest.Cell Host Microbe. 2008 Aug 14;4(2):89-91. doi: 10.1016/j.chom.2008.07.010. Cell Host Microbe. 2008. PMID: 18692766
References
-
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–1290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

